Molecular mechanism of phosphopeptide neoantigen immunogenicity
- PMID: 37353482
- PMCID: PMC10290117
- DOI: 10.1038/s41467-023-39425-1
Molecular mechanism of phosphopeptide neoantigen immunogenicity
Erratum in
-
Author Correction: Molecular mechanism of phosphopeptide neoantigen immunogenicity.Nat Commun. 2023 Jul 27;14(1):4522. doi: 10.1038/s41467-023-40274-1. Nat Commun. 2023. PMID: 37500629 Free PMC article. No abstract available.
Abstract
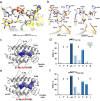
Altered protein phosphorylation in cancer cells often leads to surface presentation of phosphopeptide neoantigens. However, their role in cancer immunogenicity remains unclear. Here we describe a mechanism by which an HLA-B*0702-specific acute myeloid leukemia phosphoneoantigen, pMLL747-755 (EPR(pS)PSHSM), is recognized by a cognate T cell receptor named TCR27, a candidate for cancer immunotherapy. We show that the replacement of phosphoserine P4 with serine or phosphomimetics does not affect pMHC conformation or peptide-MHC affinity but abrogates TCR27-dependent T cell activation and weakens binding between TCR27 and pMHC. Here we describe the crystal structures for TCR27 and cognate pMHC, map of the interface produced by nuclear magnetic resonance, and a ternary complex generated using information-driven protein docking. Our data show that non-covalent interactions between the epitope phosphate group and TCR27 are crucial for TCR specificity. This study supports development of new treatment options for cancer patients through target expansion and TCR optimization.
© 2023. The Author(s).
Conflict of interest statement
All authors have directly participated in the planning, execution, or analysis of the study and drafting or revising of the manuscript, and have read and approved the final version of the manuscript. Y.P., L.P., S.N., A.N. and M.K. have no financial interests to disclose. D.U., B.M., X.M., B.J., C.B., O.H., S.G., E.C, A.S.Y., F.S., R.B.S., M.A.F., A.H., D.P., M.v.D., J.S.B. have ownership of equity securities and/or are currently or previously employed by Agenus. This does not alter our adherence to policy of the journal on sharing the data and materials.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

