Characterization of the pH-dependent protein stability of 3α-hydroxysteroid dehydrogenase/carbonyl reductase by differential scanning fluorimetry
- PMID: 37354013
- PMCID: PMC10357940
- DOI: 10.1002/pro.4710
Characterization of the pH-dependent protein stability of 3α-hydroxysteroid dehydrogenase/carbonyl reductase by differential scanning fluorimetry
Abstract
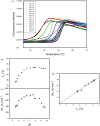
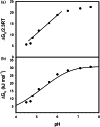
The characterization of protein stability is essential for understanding the functions of proteins. Hydroxysteroid dehydrogenase is involved in the biosynthesis of steroid hormones and the detoxification of xenobiotic carbonyl compounds. However, the stability of hydroxysteroid dehydrogenases has not yet been characterized in detail. Here, we determined the changes in Gibbs free energy, enthalpy, entropy, and heat capacity of unfolding for 3α-hydroxysteroid dehydrogenase/carbonyl reductase (3α-HSD/CR) by varying the pH and urea concentration through differential scanning fluorimetry and presented pH-dependent protein stability as a function of temperature. 3α-HSD/CR shows the maximum stability of 30.79 kJ mol-1 at 26.4°C, pH 7.6 and decreases to 7.74 kJ mol-1 at 25.7°C, pH 4.5. The change of heat capacity of 30.25 ± 1.38 kJ mol-1 K-1 is obtained from the enthalpy of denaturation as a function of melting temperature at varied pH. Two proton uptakes are linked to protein unfolding from residues with differential pKa of 4.0 and 6.5 in the native and denatured states, respectively. The large positive heat capacity change indicated that hydrophobic interactions played an important role in the folding of 3α-HSD/CR. These studies reveal the mechanism of protein unfolding in HSD and provide a convenient method to extract thermodynamic parameters for characterizing protein stability using differential scanning fluorimetry.
Keywords: Gibbs free energy; denaturant; differential scanning fluorimetry; enthalpy; entropy; heat capacity; protein stability curve; thermal unfolding.
© 2023 The Protein Society.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Contribution of remote substrate binding energy to the enzymatic rate acceleration for 3α-hydroxysteroid dehydrogenase/carbonyl reductase.Chem Biol Interact. 2017 Oct 1;276:133-140. doi: 10.1016/j.cbi.2017.01.016. Epub 2017 Jan 28. Chem Biol Interact. 2017. PMID: 28137513
-
Thermodynamic analysis of remote substrate binding energy in 3α-hydroxysteroid dehydrogenase/carbonyl reductase catalysis.Chem Biol Interact. 2019 Apr 1;302:183-189. doi: 10.1016/j.cbi.2019.02.011. Epub 2019 Feb 20. Chem Biol Interact. 2019. PMID: 30794798
-
Mechanistic roles of Ser-114, Tyr-155, and Lys-159 in 3alpha-hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni.J Biol Chem. 2005 Feb 4;280(5):3522-8. doi: 10.1074/jbc.M411751200. Epub 2004 Nov 29. J Biol Chem. 2005. PMID: 15572373
-
Protein Unfolding-Thermodynamic Perspectives and Unfolding Models.Int J Mol Sci. 2023 Mar 13;24(6):5457. doi: 10.3390/ijms24065457. Int J Mol Sci. 2023. PMID: 36982534 Free PMC article. Review.
-
Structure-function relationships in 3alpha-hydroxysteroid dehydrogenases: a comparison of the rat and human isoforms.J Steroid Biochem Mol Biol. 2003 Jun;85(2-5):247-55. doi: 10.1016/s0960-0760(03)00236-x. J Steroid Biochem Mol Biol. 2003. PMID: 12943710 Review.
Cited by
-
Measuring the Thermal Unfolding of Lysozyme: A Critical Comparison of Differential Scanning Fluorimetry and Differential Scanning Calorimetry.ChemistryOpen. 2025 Jun;14(6):e202400340. doi: 10.1002/open.202400340. Epub 2025 Feb 11. ChemistryOpen. 2025. PMID: 39935040 Free PMC article.
References
-
- Becktel WJ, Schellman JA. Protein stability curves. Biopolymers. 1987;26:1859–77. - PubMed
-
- Bergsdorf C, Wright SK. A guide to run affinity screens using differential scanning fluorimetry and surface Plasmon resonance assays. Methods Enzymol. 2018;610:135–65. - PubMed
-
- Bruylants G, Wouters J, Michaux C. Differential scanning calorimetry in life science: thermodynamics, stability, molecular recognition and application in drug design. Curr Med Chem. 2005;12:2011–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

