Copy number of naked DNA delivered into nucleus of mammalian cells by electrotransfection
- PMID: 37356265
- PMCID: PMC10527462
- DOI: 10.1016/j.bioelechem.2023.108491
Copy number of naked DNA delivered into nucleus of mammalian cells by electrotransfection
Abstract
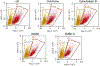
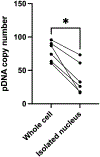
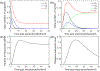
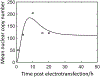
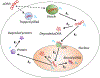
Electrotransfection is a non-viral method for delivery of nucleic acids into cells. In our previous study, we have determined the minimal copy number of plasmid DNA (pDNA) per cell required for transgene expression post electrotransfection, and developed a statistical framework to predict the pDNA copy number in the nucleus. To experimentally verify the prediction, the current study was designed to quantify the average copy number of pDNA per nucleus post electrotransfection. To achieve it, we developed a novel approach to effectively obtain isolated nuclei with minimal contamination by extranuclear pDNA. This sample preparation method enabled us to accurately measure intranuclear pDNA using quantitative real-time PCR. The data showed that the copy number of pDNA per nucleus was dependent on the period of cell culture post pulsing and the pDNA dose for electrotransfection. Additionally, the data were used to improve the statistical framework for understanding kinetics of pDNA transport in cells, and predicting how the kinetics depended on different factors. It is expected that the framework and the methodology developed in the current study will be useful for evaluating factors that may affect kinetics and mechanisms of pDNA transport in cells.
Keywords: Copy number of DNA; Electroporation; Electrotransfection; Mathematical modeling; Non-viral gene delivery; Nucleus isolation.
Copyright © 2023 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
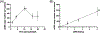
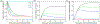
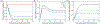
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

