Global Run-On sequencing to measure nascent transcription in Saccharomyces cerevisiae
- PMID: 37356780
- PMCID: PMC10529987
- DOI: 10.1016/j.ymeth.2023.06.009
Global Run-On sequencing to measure nascent transcription in Saccharomyces cerevisiae
Abstract
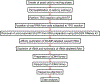
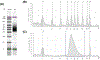
Global Run-On sequencing is a reliable and widely used approach for monitoring nascent transcription on a genomewide scale. The assay has been successfully used for studying global transcription in humans, plants, worms, flies, and fission yeast. Here we describe a GRO-seq protocol for studying transcription in budding yeast, Saccharomyces cerevisiae. Briefly, the technique involves permeabilization of actively growing yeast cells, allowing transcription to proceed in permeabilized cells in the presence of brominated UTP, affinity purification of bromo-UMP incorporated nascent transcripts followed by cDNA library construction, deep sequencing, and mapping against the reference genome. The approach maps the position of transcriptionally active RNA polymerase on a genomewide basis. In addition to identifying the complete set of transcriptionally active genes in a cell under a given set of conditions, the method can be used to determine elongation rate, termination defect and promoter directionality at the genomewide level. The approach is especially useful in identifying short-lived unstable transcripts that are rapidly degraded even before they leave the nucleus.
Keywords: Nascent RNA; RNA polymerase; Sequencing; Transcription Run-On; Transcription directionality; Transcription initiation; Transcription termination.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Liu Y, Beyer A and Aebersold R, 2016. On the dependency of cellular protein levels on mRNA abundance. Cell, 165(3), pp.535–550. - PubMed
-
- Gatehouse J and Thompson AJ, 1995. Nuclear “run-on” transcription assays. In Plant Gene Transfer and Expression Protocols (pp. 229–238). Springer, Totowa, NJ. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

