Transcriptional and post-translational regulation of plant autophagy
- PMID: 37358252
- PMCID: PMC10575704
- DOI: 10.1093/jxb/erad211
Transcriptional and post-translational regulation of plant autophagy
Abstract
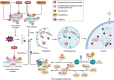
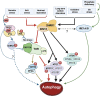
In response to changing environmental conditions, plants activate cellular responses to enable them to adapt. One such response is autophagy, in which cellular components, for example proteins and organelles, are delivered to the vacuole for degradation. Autophagy is activated by a wide range of conditions, and the regulatory pathways controlling this activation are now being elucidated. However, key aspects of how these factors may function together to properly modulate autophagy in response to specific internal or external signals are yet to be discovered. In this review we discuss mechanisms for regulation of autophagy in response to environmental stress and disruptions in cell homeostasis. These pathways include post-translational modification of proteins required for autophagy activation and progression, control of protein stability of the autophagy machinery, and transcriptional regulation, resulting in changes in transcription of genes involved in autophagy. In particular, we highlight potential connections between the roles of key regulators and explore gaps in research, the filling of which can further our understanding of the autophagy regulatory network in plants.
Keywords: ATG; autophagy; gene expression; persulfidation; phosphorylation; post-translational modification; starvation; stress; transcription factors; ubiquitination.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Conflict of interest statement
The authors declare they have no conflict of interest.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

