FAM46C Is an Interferon-Stimulated Gene That Inhibits Lentiviral Particle Production by Modulating Autophagy
- PMID: 37358411
- PMCID: PMC10434054
- DOI: 10.1128/spectrum.05211-22
FAM46C Is an Interferon-Stimulated Gene That Inhibits Lentiviral Particle Production by Modulating Autophagy
Abstract
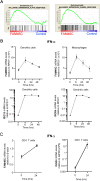
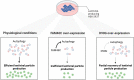
FAM46C is a multiple myeloma (MM) tumor suppressor whose function is only starting to be elucidated. We recently showed that in MM cells FAM46C triggers apoptosis by inhibiting autophagy and altering intracellular trafficking and protein secretion. To date, both a physiological characterization of FAM46C role and an assessment of FAM46C-induced phenotypes outside of MM are lacking. Preliminary reports suggested an involvement of FAM46C with regulation of viral replication, but this was never confirmed. Here, we show that FAM46C is an interferon-stimulated gene and that the expression of wild-type FAM46C in HEK-293T cells, but not of its most frequently found mutant variants, inhibits the production of both HIV-1-derived and HIV-1 lentiviruses. We demonstrate that this effect does not require transcriptional regulation and does not depend on inhibition of either global or virus-specific translation but rather mostly relies on FAM46C-induced deregulation of autophagy, a pathway that we show to be required for efficient lentiviral particle production. These studies not only provide new insights on the physiological role of the FAM46C protein but also could help in implementing more efficient antiviral strategies on one side and lentiviral particle production approaches on the other. IMPORTANCE FAM46C role has been thoroughly investigated in MM, but studies characterizing its role outside of the tumoral environment are still lacking. Despite the success of antiretroviral therapy in suppressing HIV load to undetectable levels, there is currently no HIV cure, and treatment is lifelong. Indeed, HIV continues to be a major global public health issue. Here, we show that FAM46C expression in HEK-293T cells inhibits the production of both HIV and HIV-derived lentiviruses. We also demonstrate that such inhibitory effect relies, at least in part, on the well-established regulatory role that FAM46C exerts on autophagy. Deciphering the molecular mechanism underlying this regulation will not only facilitate the understanding of FAM46C physiological role but also give new insights on the interplay between HIV and the cellular environment.
Keywords: FAM46C; HIV-1; TENT5C; autophagy; interferons; lentiviruses; viral production.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, Dawson KJ, Iorio F, Nik-Zainal S, Bignell GR, Hinton JW, Li Y, Tubio JMC, McLaren S, O’ Meara S, Butler AP, Teague JW, Mudie L, Anderson E, Rashid N, Tai Y-T, Shammas MA, Sperling AS, Fulciniti M, Richardson PG, Parmigiani G, Magrangeas F, Minvielle S, Moreau P, Attal M, Facon T, Futreal PA, Anderson KC, Campbell PJ, Munshi NC. 2014. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun 5:2997. doi:10.1038/ncomms3997. - DOI - PMC - PubMed
-
- Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, Sougnez C, Knoechel B, Gould J, Saksena G, Cibulskis K, McKenna A, Chapman MA, Straussman R, Levy J, Perkins LM, Keats JJ, Schumacher SE, Rosenberg M, Getz G, Golub TR, Anderson KC, Richardson P, Krishnan A, Lonial S, Kaufman J, Siegel DS, Vesole DH, Roy V, Rivera CE, Rajkumar SV, Kumar S, Fonseca R, Ahmann GJ, Bergsagel PL, Stewart AK, Hofmeister CC, Efebera YA, Jagannath S, Chari A, Trudel S, Reece D, Wolf J, Martin T, Zimmerman T, Rosenbaum C, Jakubowiak AJ, Lebovic D, Vij R, Stockerl-Goldstein K, Multiple Myeloma Research Consortium . 2014. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 25:91–101. doi:10.1016/j.ccr.2013.12.015. - DOI - PMC - PubMed
-
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet J-P, Ahmann GJ, Adli M, Anderson KC, Ardlie KG, Auclair D, Baker A, Bergsagel PL, Bernstein BE, Drier Y, Fonseca R, Gabriel SB, Hofmeister CC, Jagannath S, Jakubowiak AJ, Krishnan A, Levy J, Liefeld T, Lonial S, Mahan S, Mfuko B, Monti S, Perkins LM, Onofrio R, Pugh TJ, Rajkumar SV, Ramos AH, Siegel DS, Sivachenko A, Stewart AK, Trudel S, Vij R, Voet D, Winckler W, Zimmerman T, Carpten J, Trent J, Hahn WC, Garraway LA, Meyerson M, Lander ES, Getz G, Golub TR. 2011. Initial genome sequencing and analysis of multiple myeloma. Nature 471:467–472. doi:10.1038/nature09837. - DOI - PMC - PubMed
-
- Boyd KD, Ross FM, Walker BA, Wardell CP, Tapper WJ, Chiecchio L, Dagrada G, Konn ZJ, Gregory WM, Jackson GH, Child JA, Davies FE, Morgan GJ, NCRI Haematology Oncology Studies Group . 2011. Mapping of chromosome 1p deletions in myeloma identifies FAM46C at 1p12 and CDKN2C at 1p32.3 as being genes in regions associated with adverse survival. Clin Cancer Res 17:7776–7784. doi:10.1158/1078-0432.CCR-11-1791. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

