Pan-Genome-Wide Association Study of Serotype 19A Pneumococci Identifies Disease-Associated Genes
- PMID: 37358412
- PMCID: PMC10433855
- DOI: 10.1128/spectrum.04073-22
Pan-Genome-Wide Association Study of Serotype 19A Pneumococci Identifies Disease-Associated Genes
Abstract
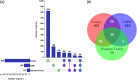
Despite the widespread implementation of pneumococcal vaccines, hypervirulent Streptococcus pneumoniae serotype 19A is endemic worldwide. It is still unclear whether specific genetic elements contribute to complex pathogenicity of serotype 19A isolates. We performed a large-scale pan-genome-wide association study (pan-GWAS) of 1,292 serotype 19A isolates sampled from patients with invasive disease and asymptomatic carriers. To address the underlying disease-associated genotypes, a comprehensive analysis using three methods (Scoary, a linear mixed model, and random forest) was performed to compare disease and carriage isolates to identify genes consistently associated with disease phenotype. By using three pan-GWAS methods, we found consensus on statistically significant associations between genotypes and disease phenotypes (disease or carriage), with a subset of 30 consistently significant disease-associated genes. The results of functional annotation revealed that these disease-associated genes had diverse predicted functions, including those that participated in mobile genetic elements, antibiotic resistance, virulence, and cellular metabolism. Our findings suggest the multifactorial pathogenicity nature of this hypervirulent serotype and provide important evidence for the design of novel protein-based vaccines to prevent and control pneumococcal disease. IMPORTANCE It is important to understand the genetic and pathogenic characteristics of S. pneumoniae serotype 19A, which may provide important information for the prevention and treatment of pneumococcal disease. This global large-sample pan-GWAS study has identified a subset of 30 consistently significant disease-associated genes that are involved in mobile genetic elements, antibiotic resistance, virulence, and cellular metabolism. These findings suggest the multifactorial pathogenicity nature of hypervirulent S. pneumoniae serotype 19A isolates and provide implications for the design of novel protein-based vaccines.
Keywords: Streptococcus pneumoniae; bacterial genomics; carriage; genome-wide association study; invasive pneumococcal disease; pathogenicity; pneumococcus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
CIRCULATING CLONAL COMPLEXES AND SEQUENCE TYPES OF STREPTOCOCCUS PNEUMONIAE SEROTYPE 19A WORLDWIDE: THE IMPORTANCE OF MULTIDRUG RESISTANCE: A SYSTEMATIC LITERATURE REVIEW.Expert Rev Vaccines. 2021 Jan;20(1):45-57. doi: 10.1080/14760584.2021.1873136. Epub 2021 Feb 17. Expert Rev Vaccines. 2021. PMID: 33507135
-
Using genomics to examine the persistence of Streptococcus pneumoniae serotype 19A in Ireland and the emergence of a sub-clade associated with vaccine failures.Vaccine. 2021 Aug 16;39(35):5064-5073. doi: 10.1016/j.vaccine.2021.06.017. Epub 2021 Jul 21. Vaccine. 2021. PMID: 34301430
-
Pneumococcal conjugate vaccination and nasopharyngeal acquisition of pneumococcal serotype 19A strains.JAMA. 2010 Sep 8;304(10):1099-106. doi: 10.1001/jama.2010.1290. JAMA. 2010. PMID: 20823436
-
Characteristics of Streptococcus pneumoniae serotype 19A isolates from children in the pre and post Conjugate Vaccine Era. Single center experience 1986-2015.Vaccine. 2018 Aug 23;36(35):5245-5250. doi: 10.1016/j.vaccine.2018.07.055. Epub 2018 Jul 27. Vaccine. 2018. PMID: 30061027
-
Streptococcus pneumoniae serotype 19A: worldwide epidemiology.Expert Rev Vaccines. 2017 Oct;16(10):1007-1027. doi: 10.1080/14760584.2017.1362339. Epub 2017 Aug 28. Expert Rev Vaccines. 2017. PMID: 28783380 Review.
Cited by
-
Comparative genomic analysis of 255 Oenococcus oeni isolates from China: unveiling strain diversity and genotype-phenotype associations of acid resistance.Microbiol Spectr. 2025 Jun 3;13(6):e0326524. doi: 10.1128/spectrum.03265-24. Epub 2025 Apr 22. Microbiol Spectr. 2025. PMID: 40261018 Free PMC article.
-
Simple and accurate genomic classification model for distinguishing between human and pig Staphylococcus aureus.Commun Biol. 2024 Sep 18;7(1):1171. doi: 10.1038/s42003-024-06883-2. Commun Biol. 2024. PMID: 39294434 Free PMC article.
-
Genome-based model for differentiating between infection and carriage Staphylococcus aureus.Microbiol Spectr. 2024 Oct 3;12(10):e0049324. doi: 10.1128/spectrum.00493-24. Epub 2024 Sep 9. Microbiol Spectr. 2024. PMID: 39248515 Free PMC article.
-
Bacterial genome-wide association studies: exploring the genetic variation underlying bacterial phenotypes.Appl Environ Microbiol. 2025 Jun 18;91(6):e0251224. doi: 10.1128/aem.02512-24. Epub 2025 May 16. Appl Environ Microbiol. 2025. PMID: 40377303 Free PMC article. Review.
-
The impact of pneumococcal serotype replacement on the effectiveness of a national immunization program: a population-based active surveillance cohort study in New Zealand.Lancet Reg Health West Pac. 2024 May 8;46:101082. doi: 10.1016/j.lanwpc.2024.101082. eCollection 2024 May. Lancet Reg Health West Pac. 2024. PMID: 38745973 Free PMC article.
References
-
- Zhou M, Wang Z, Zhang L, Kudinha T, An H, Qian C, Jiang B, Wang Y, Xu Y, Liu Z, Zhang H, Zhang J. 2021. Serotype distribution, antimicrobial susceptibility, multilocus sequencing type and virulence of invasive Streptococcus pneumoniae in China: a six-year multicenter study. Front Microbiol 12:798750. doi:10.3389/fmicb.2021.798750. - DOI - PMC - PubMed
-
- Kellner JD, Ricketson LJ, Demczuk WHB, Martin I, Tyrrell GJ, Vanderkooi OG, Mulvey MR. 2021. Whole-genome analysis of Streptococcus pneumoniae serotype 4 causing outbreak of invasive pneumococcal disease, Alberta, Canada. Emerg Infect Dis 27:1867–1875. doi:10.3201/eid2707.204403. - DOI - PMC - PubMed
-
- Varghese R, Daniel JL, Neeravi A, Baskar P, Manoharan A, Sundaram B, Manchanda V, Saigal K, Yesudhasan BL, Veeraraghavan B. 2021. Multicentric analysis of erythromycin resistance determinants in invasive Streptococcus pneumoniae; associated serotypes and sequence types in India. Curr Microbiol 78:3239–3245. doi:10.1007/s00284-021-02594-7. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

