Investigating the Genetic Diversity of H5 Avian Influenza Viruses in the United Kingdom from 2020-2022
- PMID: 37358418
- PMCID: PMC10433820
- DOI: 10.1128/spectrum.04776-22
Investigating the Genetic Diversity of H5 Avian Influenza Viruses in the United Kingdom from 2020-2022
Abstract
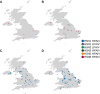
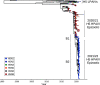
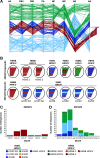
Since 2020, the United Kingdom and Europe have experienced annual epizootics of high-pathogenicity avian influenza virus (HPAIV). The first epizootic, during the autumn/winter of 2020-2021, involved six H5Nx subtypes, although H5N8 HPAIV dominated in the United Kingdom. While genetic assessments of the H5N8 HPAIVs within the United Kingdom demonstrated relative homogeneity, there was a background of other genotypes circulating at a lower degree with different neuraminidase and internal genes. Following a small number of detections of H5N1 in wild birds over the summer of 2021, the autumn/winter of 2021-2022 saw another European H5 HPAIV epizootic that dwarfed the prior epizootic. This second epizootic was dominated almost exclusively by H5N1 HPAIV, although six distinct genotypes were defined. We have used genetic analysis to evaluate the emergence of different genotypes and proposed reassortment events that have been observed. The existing data suggest that the H5N1 viruses circulating in Europe during late 2020 continued to circulate in wild birds throughout 2021, with minimal adaptation, but then went on to reassort with AIVs in the wild bird population. We have undertaken an in-depth genetic assessment of H5 HPAIVs detected in the United Kingdom over two winter seasons and demonstrate the utility of in-depth genetic analyses in defining the diversity of H5 HPAIVs circulating in avian species, the potential for zoonotic risk, and whether incidents of lateral spread can be defined over independent incursions of infections from wild birds. This provides key supporting data for mitigation activities. IMPORTANCE High-pathogenicity avian influenza virus (HPAIV) outbreaks devastate avian species across all sectors, having both economic and ecological impacts through mortalities in poultry and wild birds, respectively. These viruses can also represent a significant zoonotic risk. Since 2020, the United Kingdom has experienced two successive outbreaks of H5 HPAIV. While H5N8 HPAIV was predominant during the 2020-2021 outbreak, other H5 subtypes were also detected. The following year, there was a shift in the subtype dominance to H5N1 HPAIV, but multiple H5N1 genotypes were detected. Through the thorough utilization of whole-genome sequencing, it was possible to track and characterize the genetic evolution of these H5 HPAIVs in United Kingdom poultry and wild birds. This enabled us to assess the risk posed by these viruses at the poultry-wild bird and the avian-human interfaces and to investigate the potential lateral spread between infected premises, a key factor in understanding the threat to the commercial sector.
Keywords: WGS; avian influenza; genomics; high-pathogenicity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- WOAH. 2021. Terrestrial manual: avian influenza (including infection with high pathogenicity avian influenza viruses). WOAH, Paris, France.
-
- European Commission. 2006. Council directive 2005/94/EC of 20 December 2005 on community measures for the control of avian influenza and repealing directive 92/40/EEC. Off J Eur Union 49:L10.
-
- Vapnek J. 2010. Regulatory measures against outbreaks of highly pathogenic avian influenza. FAO Legal Papers Online #82. Food and Agriculture Organization of the United Nations, Rome, Italy.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

