Reverse Genetic Assessment of the Roles Played by the Spike Protein and ORF3 in Porcine Epidemic Diarrhea Virus Pathogenicity
- PMID: 37358450
- PMCID: PMC10373562
- DOI: 10.1128/jvi.01964-22
Reverse Genetic Assessment of the Roles Played by the Spike Protein and ORF3 in Porcine Epidemic Diarrhea Virus Pathogenicity
Abstract
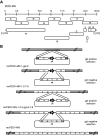
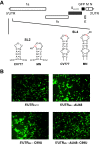
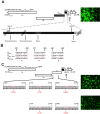
Porcine epidemic diarrhea virus is a swine pathogen that has been responsible for significant animal and economic losses worldwide in recent years. In this manuscript, we report the generation of a reverse genetics system C(RGS) for the highly virulent US PEDV strain Minnesota (PEDV-MN; GenBank accession number KF468752), which was based on the assembly and cloning of synthetic DNA, using vaccinia virus as a cloning vector. Viral rescue was only possible following the substitution of 2 nucleotides within the 5'UTR and 2 additional nucleotides within the spike gene, based on the sequence of the cell culture-adapted strains. Besides displaying a highly pathogenic phenotype in newborn piglets, in comparison with the parental virus, the rescued recombinant PEDV-MN was used to confirm that the PEDV spike gene has an important role in PEDV virulence and that the impact of an intact PEDV ORF3 on viral pathogenicity is modest. Moreover, a chimeric virus with a TGEV spike gene in the PEDV backbone generated with RGS was able to replicate efficiently in vivo and could be readily transmitted between piglets. Although this chimeric virus did not cause severe disease upon the initial infection of piglets, there was evidence of increasing pathogenicity upon transmission to contact piglets. The RGS described in this study constitutes a powerful tool with which to study PEDV pathogenesis and can be used to generate vaccines against porcine enteric coronaviruses. IMPORTANCE PEDV is a swine pathogen that is responsible for significant animal and economic losses worldwide. Highly pathogenic variants can lead to a mortality rate of up to 100% in newborn piglets. The generation of a reverse genetics system for a highly virulent PEDV strain originating from the United States is an important step in phenotypically characterizing PEDV. The synthetic PEDV mirrored the authentic isolate and displayed a highly pathogenic phenotype in newborn piglets. With this system, it was possible to characterize potential viral virulence factors. Our data revealed that an accessory gene (ORF3) has a limited impact on pathogenicity. However, as it is also now known for many coronaviruses, the PEDV spike gene is one of the main determinants of pathogenicity. Finally, we show that the spike gene of another porcine coronavirus, namely, TGEV, can be accommodated in the PEDV genome background, suggesting that similar viruses can emerge in the field via recombination.
Keywords: ORF3; porcine epidemic diarrhea virus; spike gene; synthesized DNA; vaccinia virus reverse genetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Cooper VL, Pillatzki A, Gauger P, Schmitt BJ, Koster LG, Killian ML, Yoon KJ. 2013. Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest 25:649–654. doi: 10.1177/1040638713501675. - DOI - PubMed
-
- De Groot R, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, Holmes KV, Perlman S, Poon LL, Rottier P, Talbot PJ, Woo PC, Ziebuhr J. 2012. Family Coronaviridae. p 806–828. In King AMQ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, Amsterdam, NL.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

