Triglyceride-rich lipoprotein remnants, low-density lipoproteins, and risk of coronary heart disease: a UK Biobank study
- PMID: 37358553
- PMCID: PMC10576615
- DOI: 10.1093/eurheartj/ehad337
Triglyceride-rich lipoprotein remnants, low-density lipoproteins, and risk of coronary heart disease: a UK Biobank study
Abstract
Aims: The strength of the relationship of triglyceride-rich lipoproteins (TRL) with risk of coronary heart disease (CHD) compared with low-density lipoprotein (LDL) is yet to be resolved.
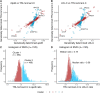
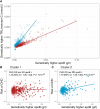
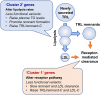
Methods and results: Single-nucleotide polymorphisms (SNPs) associated with TRL/remnant cholesterol (TRL/remnant-C) and LDL cholesterol (LDL-C) were identified in the UK Biobank population. In a multivariable Mendelian randomization analysis, TRL/remnant-C was strongly and independently associated with CHD in a model adjusted for apolipoprotein B (apoB). Likewise, in a multivariable model, TRL/remnant-C and LDL-C also exhibited independent associations with CHD with odds ratios per 1 mmol/L higher cholesterol of 2.59 [95% confidence interval (CI): 1.99-3.36] and 1.37 [95% CI: 1.27-1.48], respectively. To examine the per-particle atherogenicity of TRL/remnants and LDL, SNPs were categorized into two clusters with differing effects on TRL/remnant-C and LDL-C. Cluster 1 contained SNPs in genes related to receptor-mediated lipoprotein removal that affected LDL-C more than TRL/remnant-C, whereas cluster 2 contained SNPs in genes related to lipolysis that had a much greater effect on TRL/remnant-C. The CHD odds ratio per standard deviation (Sd) higher apoB for cluster 2 (with the higher TRL/remnant to LDL ratio) was 1.76 (95% CI: 1.58-1.96), which was significantly greater than the CHD odds ratio per Sd higher apoB in cluster 1 [1.33 (95% CI: 1.26-1.40)]. A concordant result was obtained by using polygenic scores for each cluster to relate apoB to CHD risk.
Conclusion: Distinct SNP clusters appear to impact differentially on remnant particles and LDL. Our findings are consistent with TRL/remnants having a substantially greater atherogenicity per particle than LDL.
Keywords: Apolipoprotein B; Cardiovascular disease; Genetics; LDL cholesterol; Mendelian randomization; Remnants; Single-nucleotide polymorphisms; Triglyceride; UK Biobank.
© The Author(s) 2023. Published by Oxford University Press on behalf of the European Society of Cardiology.
Figures


Comment in
-
Triglyceride-rich remnant lipoproteins are more atherogenic than LDL per particle: is this important?Eur Heart J. 2023 Oct 14;44(39):4196-4198. doi: 10.1093/eurheartj/ehad419. Eur Heart J. 2023. PMID: 37403539 No abstract available.
References
-
- Ginsberg HN, Packard CJ, Chapman MJ, Boren J, Aguilar-Salinas CA, Averna M, et al. . Triglyceride-rich lipoproteins and their remnants: metabolic insights, role in atherosclerotic cardiovascular disease, and emerging therapeutic strategies-a consensus statement from the European Atherosclerosis Society. Eur Heart J 2021;42:4791–4806. 10.1093/eurheartj/ehab551 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

