Construction of HBV-HCC prognostic model and immune characteristics based on potential genes mining through protein interaction networks
- PMID: 37358667
- PMCID: PMC11796638
- DOI: 10.1007/s00432-023-04989-4
Construction of HBV-HCC prognostic model and immune characteristics based on potential genes mining through protein interaction networks
Abstract
Objective: To search for human protein-coding genes related to hepatocellular carcinoma (HCC) in the context of hepatitis B virus (HBV) infection, and perform prognosis risk assessment.
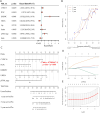
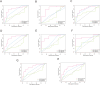
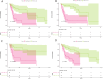
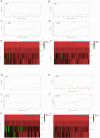
Methods: Genes related to HBV-HCC were selected through literature screening and protein-protein interaction (PPI) network database analysis. Prognosis potential genes (PPGs) were identified using Cox regression analysis. Patients were divided into high-risk and low-risk groups based on PPGs, and risk scores were calculated. Kaplan-Meier plots were used to analyze overall survival rates, and the results were predicted based on clinicopathological variables. Association analysis was also conducted with immune infiltration, immune therapy, and drug sensitivity. Experimental verification of the expression of PPGs was done in patient liver cancer tissue and normal liver tissue adjacent to tumors.
Results: The use of a prognosis potential genes risk assessment model can reliably predict the prognosis risk of patients, demonstrating strong predictive ability. Kaplan-Meier analysis showed that the overall survival rate of the low-risk group was significantly higher than that of the high-risk group. There were significant differences between the two subgroups in terms of immune infiltration and IC50 association analysis. Experimental verification revealed that CYP2C19, FLNC, and HNRNPC were highly expressed in liver cancer tissue, while UBE3A was expressed at a lower level.
Conclusion: PPGs can be used to predict the prognosis risk of HBV-HCC patients and play an important role in the diagnosis and treatment of liver cancer. They also reveal their potential role in the tumor immune microenvironment, clinical-pathological characteristics, and prognosis.
Keywords: Chemotherapy sensitivity; HBV-HCC; Immune infiltration; PPI; Prognosis.
© 2023. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
A novel prognostic model based on immunogenic cell death-related genes for improved risk stratification in hepatocellular carcinoma patients.J Cancer Res Clin Oncol. 2023 Sep;149(12):10255-10267. doi: 10.1007/s00432-023-04950-5. Epub 2023 Jun 3. J Cancer Res Clin Oncol. 2023. PMID: 37269346 Free PMC article.
-
Prediction of the effect of Bupleurum chinensis DC. (Chaihu) on survival and its potential targets in hepatocellular carcinoma based on a prognostic model.BMC Complement Med Ther. 2025 Jul 3;25(1):228. doi: 10.1186/s12906-025-04848-4. BMC Complement Med Ther. 2025. PMID: 40611075 Free PMC article.
-
Pharmacological interventions for acute hepatitis B infection: an attempted network meta-analysis.Cochrane Database Syst Rev. 2017 Mar 21;3(3):CD011645. doi: 10.1002/14651858.CD011645.pub2. Cochrane Database Syst Rev. 2017. PMID: 28321877 Free PMC article.
-
Nuclear factor IA-mediated transcriptional regulation of crystallin αB inhibits hepatocellular carcinoma progression.Mol Clin Oncol. 2025 Jun 20;23(2):72. doi: 10.3892/mco.2025.2867. eCollection 2025 Aug. Mol Clin Oncol. 2025. PMID: 40599718 Free PMC article.
-
Adefovir dipivoxil and pegylated interferon alfa-2a for the treatment of chronic hepatitis B: a systematic review and economic evaluation.Health Technol Assess. 2006 Aug;10(28):iii-iv, xi-xiv, 1-183. doi: 10.3310/hta10280. Health Technol Assess. 2006. PMID: 16904047
References
-
- Abdelgalil AA, Alkahtani HM, Al-Jenoobi FI (2019) Sorafenib. Profiles Drug Subst Excip Relat Methodol 44:239–266 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

