Molecular crosstalk between COVID-19 and Alzheimer's disease using microarray and RNA-seq datasets: A system biology approach
- PMID: 37359008
- PMCID: PMC10286240
- DOI: 10.3389/fmed.2023.1151046
Molecular crosstalk between COVID-19 and Alzheimer's disease using microarray and RNA-seq datasets: A system biology approach
Abstract
Objective: Coronavirus disease 2019 (COVID-19) is an infectious disease caused by Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2). The clinical and epidemiological analysis reported the association between SARS-CoV-2 and neurological diseases. Among neurological diseases, Alzheimer's disease (AD) has developed as a crucial comorbidity of SARS-CoV-2. This study aimed to understand the common transcriptional signatures between SARS-CoV-2 and AD.
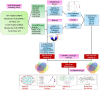
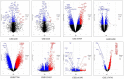
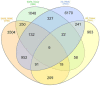
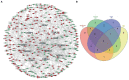
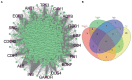
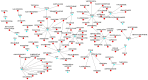
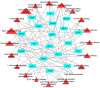
Materials and methods: System biology approaches were used to compare the datasets of AD and COVID-19 to identify the genetic association. For this, we have integrated three human whole transcriptomic datasets for COVID-19 and five microarray datasets for AD. We have identified differentially expressed genes for all the datasets and constructed a protein-protein interaction (PPI) network. Hub genes were identified from the PPI network, and hub genes-associated regulatory molecules (transcription factors and miRNAs) were identified for further validation.
Results: A total of 9,500 differentially expressed genes (DEGs) were identified for AD and 7,000 DEGs for COVID-19. Gene ontology analysis resulted in 37 molecular functions, 79 cellular components, and 129 biological processes were found to be commonly enriched in AD and COVID-19. We identified 26 hub genes which includes AKT1, ALB, BDNF, CD4, CDH1, DLG4, EGF, EGFR, FN1, GAPDH, INS, ITGB1, ACTB, SRC, TP53, CDC42, RUNX2, HSPA8, PSMD2, GFAP, VAMP2, MAPK8, CAV1, GNB1, RBX1, and ITGA2B. Specific miRNA targets associated with Alzheimer's disease and COVID-19 were identified through miRNA target prediction. In addition, we found hub genes-transcription factor and hub genes-drugs interaction. We also performed pathway analysis for the hub genes and found that several cell signaling pathways are enriched, such as PI3K-AKT, Neurotrophin, Rap1, Ras, and JAK-STAT.
Conclusion: Our results suggest that the identified hub genes could be diagnostic biomarkers and potential therapeutic drug targets for COVID-19 patients with AD comorbidity.
Keywords: Alzheimer’s disease; COVID-19; biomarkers; comorbidity; regulatory networks.
Copyright © 2023 Premkumar and Sajitha Lulu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Analysis and Identification Genetic Effect of SARS-CoV-2 Infections to Alzheimer's Disease Patients by Integrated Bioinformatics.J Alzheimers Dis. 2022;85(2):729-744. doi: 10.3233/JAD-215086. J Alzheimers Dis. 2022. PMID: 34776447
-
Unraveling the molecular crosstalk and immune landscape between COVID-19 infections and ischemic heart failure comorbidity: New insights into diagnostic biomarkers and therapeutic approaches.Cell Signal. 2023 Dec;112:110909. doi: 10.1016/j.cellsig.2023.110909. Epub 2023 Sep 28. Cell Signal. 2023. PMID: 37777104
-
Identification of common molecular signatures of SARS-CoV-2 infection and its influence on acute kidney injury and chronic kidney disease.Front Immunol. 2023 Mar 21;14:961642. doi: 10.3389/fimmu.2023.961642. eCollection 2023. Front Immunol. 2023. PMID: 37026010 Free PMC article.
-
The interference between SARS-COV-2 and Alzheimer's disease: Potential immunological and neurobiological crosstalk from a kinase perspective reveals a delayed pandemic.Ageing Res Rev. 2024 Feb;94:102195. doi: 10.1016/j.arr.2024.102195. Epub 2024 Jan 18. Ageing Res Rev. 2024. PMID: 38244862 Review.
-
COVID-19 and Alzheimer's Disease.Brain Sci. 2021 Feb 27;11(3):305. doi: 10.3390/brainsci11030305. Brain Sci. 2021. PMID: 33673697 Free PMC article. Review.
Cited by
-
Caveolin-1 mediates blood-brain barrier permeability, neuroinflammation, and cognitive impairment in SARS-CoV-2 infection.J Neuroimmunol. 2024 Mar 15;388:578309. doi: 10.1016/j.jneuroim.2024.578309. Epub 2024 Feb 4. J Neuroimmunol. 2024. PMID: 38335781 Free PMC article.
-
Unveiling reverse vaccinology and immunoinformatics toward Saint Louis encephalitis virus: a ray of hope for vaccine development.Front Immunol. 2025 May 19;16:1576557. doi: 10.3389/fimmu.2025.1576557. eCollection 2025. Front Immunol. 2025. PMID: 40458397 Free PMC article.
-
Cell-specific transcriptional signatures of vascular cells in Alzheimer's disease: perspectives, pathways, and therapeutic directions.Mol Neurodegener. 2025 Jan 29;20(1):12. doi: 10.1186/s13024-025-00798-0. Mol Neurodegener. 2025. PMID: 39876020 Free PMC article. Review.
-
Alzheimer's disease and infectious agents: a comprehensive review of pathogenic mechanisms and microRNA roles.Front Neurosci. 2025 Jan 7;18:1513095. doi: 10.3389/fnins.2024.1513095. eCollection 2024. Front Neurosci. 2025. PMID: 39840010 Free PMC article. Review.
-
Parallel electrophysiological abnormalities due to COVID-19 infection and to Alzheimer's disease and related dementia.Alzheimers Dement. 2024 Oct;20(10):7296-7319. doi: 10.1002/alz.14089. Epub 2024 Aug 29. Alzheimers Dement. 2024. PMID: 39206795 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

