Integrative Analysis of Proteomics and Transcriptomics Reveals Endothelin Receptor B as Novel Single Target and Identifies New Combinatorial Targets for Multiple Myeloma
- PMID: 37359190
- PMCID: PMC10289631
- DOI: 10.1097/HS9.0000000000000901
Integrative Analysis of Proteomics and Transcriptomics Reveals Endothelin Receptor B as Novel Single Target and Identifies New Combinatorial Targets for Multiple Myeloma
Abstract
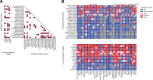
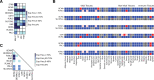
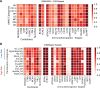
Despite the recent introduction of next-generation immunotherapeutic agents, multiple myeloma (MM) remains incurable. New strategies targeting MM-specific antigens may result in a more effective therapy by preventing antigen escape, clonal evolution, and tumor resistance. In this work, we adapted an algorithm that integrates proteomic and transcriptomic results of myeloma cells to identify new antigens and possible antigen combinations. We performed cell surface proteomics on 6 myeloma cell lines based and combined these results with gene expression studies. Our algorithm identified 209 overexpressed surface proteins from which 23 proteins could be selected for combinatorial pairing. Flow cytometry analysis of 20 primary samples confirmed the expression of FCRL5, BCMA, and ICAM2 in all samples and IL6R, endothelin receptor B (ETB), and SLCO5A1 in >60% of myeloma cases. Analyzing possible combinations, we found 6 combinatorial pairs that can target myeloma cells and avoid toxicity on other organs. In addition, our studies identified ETB as a tumor-associated antigen that is overexpressed on myeloma cells. This antigen can be targeted with a new monoclonal antibody RB49 that recognizes an epitope located in a region that becomes highly accessible after activation of ETB by its ligand. In conclusion, our algorithm identified several candidate antigens that can be used for either single-antigen targeting approaches or for combinatorial targeting in new immunotherapeutic approaches in MM.
Copyright © 2023 the Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the European Hematology Association.
Conflict of interest statement
DB and AH are scientific cofounders and hold equity in Skymab Biotherapeutics. All the other authors have no conflicts of interest to disclose.
Figures



References
-
- Lonial S, Weiss BM, Usmani SZ, et al. . Daratumumab monotherapy in patients with treatment-refractory multiple myeloma (SIRIUS): an open-label, randomised, phase 2 trial. Lancet. 2016;387:1551–1560. - PubMed
-
- Dimopoulos MA, Oriol A, Nahi H, et al. . Daratumumab, lenalidomide, and dexamethasone for multiple myeloma. N Engl J Med. 2016;375:1319–1331. - PubMed
-
- Palumbo A, Chanan-Khan A, Weisel K, et al. . Daratumumab, bortezomib, and dexamethasone for multiple myeloma. N Engl J Med. 2016;375:754–766. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
