Engineering B cells with customized therapeutic responses using a synthetic circuit
- PMID: 37359346
- PMCID: PMC10285500
- DOI: 10.1016/j.omtn.2023.05.024
Engineering B cells with customized therapeutic responses using a synthetic circuit
Abstract
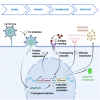
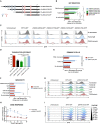
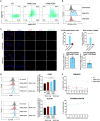
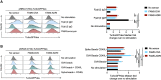
The expansion of genetic engineering has brought a new dimension for synthetic immunology. Immune cells are perfect candidates because of their ability to patrol the body, interact with many cell types, proliferate upon activation, and differentiate in memory cells. This study aimed at implementing a new synthetic circuit in B cells, allowing the expression of therapeutic molecules in a temporally and spatially restricted manner that is induced by the presence of specific antigens. This should enhance endogenous B cell functions in terms of recognition and effector properties. We developed a synthetic circuit encoding a sensor (a membrane-anchored B cell receptor targeting a model antigen), a transducer (a minimal promoter induced by the activated sensor), and effector molecules. We isolated a 734-bp-long fragment of the NR4A1 promoter, specifically activated by the sensor signaling cascade in a fully reversible manner. We demonstrate full antigen-specific circuit activation as its recognition by the sensor induced the activation of the NR4A1 promoter and the expression of the effector. Overall, such novel synthetic circuits offer huge possibilities for the treatment of many pathologies, as they are completely programmable; thus, the signal-specific sensors and effector molecules can be adapted to each disease.
Keywords: B cells; MT: Delivery Strategies; autologous; cell therapy; personalized medicine; reprogramming; synthetic circuits.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials

