Genomics reveal the origins and current structure of a genetically depauperate freshwater species in its introduced Alaskan range
- PMID: 37360023
- PMCID: PMC10286226
- DOI: 10.1111/eva.13556
Genomics reveal the origins and current structure of a genetically depauperate freshwater species in its introduced Alaskan range
Abstract
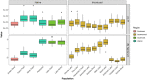
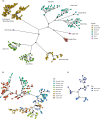
Invasive species are a major threat to global biodiversity, yet also represent large-scale unplanned ecological and evolutionary experiments to address fundamental questions in nature. Here we analyzed both native and invasive populations of predatory northern pike (Esox lucius) to characterize landscape genetic variation, determine the most likely origins of introduced populations, and investigate a presumably postglacial population from Southeast Alaska of unclear provenance. Using a set of 4329 SNPs from 351 individual Alaskan northern pike representing the most widespread geographic sampling to date, our results confirm low levels of genetic diversity in native populations (average 𝝅 of 3.18 × 10-4) and even less in invasive populations (average 𝝅 of 2.68 × 10-4) consistent with bottleneck effects. Our analyses indicate that invasive northern pike likely came from multiple introductions from different native Alaskan populations and subsequently dispersed from original introduction sites. At the broadest scale, invasive populations appear to have been founded from two distinct regions of Alaska, indicative of two independent introduction events. Genetic admixture resulting from introductions from multiple source populations may have mitigated the negative effects associated with genetic bottlenecks in this species with naturally low levels of genetic diversity. Genomic signatures strongly suggest an excess of rare, population-specific alleles, pointing to a small number of founding individuals in both native and introduced populations consistent with a species' life history of limited dispersal and gene flow. Lastly, the results strongly suggest that a small isolated population of pike, located in Southeast Alaska, is native in origin rather than stemming from a contemporary introduction event. Although theory predicts that lack of genetic variation may limit colonization success of novel environments, we detected no evidence that a lack of standing variation limited the success of this genetically depauperate apex predator.
Keywords: Esox lucius; dispersal; gene flow; invasive species; northern pike; population genetics.
© 2023 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no competing interests to declare.
Figures






References
-
- Andrews, S. , Krueger, F. , Segonds‐Pichon, A. , Biggins, L. , Krueger, C. , & Wingett, S. (2010). FastQC. A quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Beerli, P. (2004). Effect of unsampled populations on the estimation of population sizes and migration rates between sampled populations. Molecular Ecology, 13(4), 827–836. - PubMed
-
- Berghaus, K. I. , Spencer, J. R. , & Westley, P. A. (2019). Contemporary phenotypic divergence of an introduced predatory freshwater fish, the northern pike (Esox lucius). Evolutionary Ecology Research, 20(5), 487–504.
-
- Carim, K. J. , Caleb Dysthe, J. , McLellan, H. , Young, M. K. , McKelvey, K. S. , & Schwartz, M. K. (2019). Using environmental DNA sampling to monitor the invasion of nonnative Esox lucius (northern pike) in the Columbia River basin, USA. Environmental DNA, 1(3), 215–226.
-
- Carim, K. J. , Eby, L. A. , Miller, L. M. , McLellan, H. , Dupuis, V. , & Schwartz, M. K. (2022). Mechanism of northern pikeinvasion in the Columbia River Basin. Management of Biological Invasions, 13(1), 168–190. 10.3391/mbi.2022.13.1.10 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

