More than mcr: canonical plasmid- and transposon-encoded mobilized colistin resistance genes represent a subset of phosphoethanolamine transferases
- PMID: 37360531
- PMCID: PMC10285318
- DOI: 10.3389/fcimb.2023.1060519
More than mcr: canonical plasmid- and transposon-encoded mobilized colistin resistance genes represent a subset of phosphoethanolamine transferases
Abstract
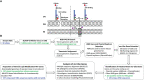
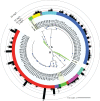
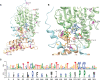
Mobilized colistin resistance genes (mcr) may confer resistance to the last-resort antimicrobial colistin and can often be transmitted horizontally. mcr encode phosphoethanolamine transferases (PET), which are closely related to chromosomally encoded, intrinsic lipid modification PET (i-PET; e.g., EptA, EptB, CptA). To gain insight into the evolution of mcr within the context of i-PET, we identified 69,814 MCR-like proteins present across 256 bacterial genera (obtained by querying known MCR family representatives against the National Center for Biotechnology Information [NCBI] non-redundant protein database via protein BLAST). We subsequently identified 125 putative novel mcr-like genes, which were located on the same contig as (i) ≥1 plasmid replicon and (ii) ≥1 additional antimicrobial resistance gene (obtained by querying the PlasmidFinder database and NCBI's National Database of Antibiotic Resistant Organisms, respectively, via nucleotide BLAST). At 80% amino acid identity, these putative novel MCR-like proteins formed 13 clusters, five of which represented putative novel MCR families. Sequence similarity and a maximum likelihood phylogeny of mcr, putative novel mcr-like, and ipet genes indicated that sequence similarity was insufficient to discriminate mcr from ipet genes. A mixed-effect model of evolution (MEME) indicated that site- and branch-specific positive selection played a role in the evolution of alleles within the mcr-2 and mcr-9 families. MEME suggested that positive selection played a role in the diversification of several residues in structurally important regions, including (i) a bridging region that connects the membrane-bound and catalytic periplasmic domains, and (ii) a periplasmic loop juxtaposing the substrate entry tunnel. Moreover, eptA and mcr were localized within different genomic contexts. Canonical eptA genes were typically chromosomally encoded in an operon with a two-component regulatory system or adjacent to a TetR-type regulator. Conversely, mcr were represented by single-gene operons or adjacent to pap2 and dgkA, which encode a PAP2 family lipid A phosphatase and diacylglycerol kinase, respectively. Our data suggest that eptA can give rise to "colistin resistance genes" through various mechanisms, including mobilization, selection, and diversification of genomic context and regulatory pathways. These mechanisms likely altered gene expression levels and enzyme activity, allowing bona fide eptA to evolve to function in colistin resistance.
Keywords: MCR; antimicrobial resistance; colistin; horizontal gene transfer; mobile genetic element; phosphoethanolamine transferase; plasmid.
Copyright © 2023 Gaballa, Wiedmann and Carroll.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Abdul Momin M. H. F., Liakopoulos A., Bean D. C., Phee L. M., Wareham D. W. (2019). A novel plasmid-mediated polymyxin resistance determinant (mcr-1.8) in Escherichia coli recovered from broiler chickens in Brunei darussalam. J. Antimicrob. Chemother. 74, 3392–3394. doi: 10.1093/jac/dkz352 - DOI - PubMed
-
- Abuoun M., Stubberfield E. J., Duggett N. A., Kirchner M., Dormer L., Nunez-Garcia J., et al. (2017). mcr-1 and mcr-2 variant genes identified in Moraxella species isolated from pigs in great Britain from 2014 to 2015. J. Antimicrob. Chemother. 72, 2745–2749. doi: 10.1093/jac/dkx286 - DOI - PMC - PubMed
-
- Alba P., Leekitcharoenphon P., Franco A., Feltrin F., Ianzano A., Caprioli A., et al. (2018). Molecular epidemiology of mcr-encoded colistin resistance in enterobacteriaceae from food-producing animals in Italy revealed through the EU harmonized antimicrobial resistance monitoring. Front. Microbiol. 9, 1217. doi: 10.3389/fmicb.2018.01217 - DOI - PMC - PubMed
-
- Anandan A., Evans G. L., Condic-Jurkic K., O'mara M. L., John C. M., Phillips N. J., et al. (2017). Structure of a lipid a phosphoethanolamine transferase suggests how conformational changes govern substrate binding. Proc. Natl. Acad. Sci. U.S.A. 114, 2218–2223. doi: 10.1073/pnas.1612927114 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

