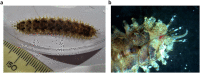
The genome sequence of the scale worm, Lepidonotus clava (Montagu, 1808)
- PMID: 37362008
- PMCID: PMC10288160
- DOI: 10.12688/wellcomeopenres.18660.1
The genome sequence of the scale worm, Lepidonotus clava (Montagu, 1808)
Abstract
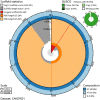
We present a genome assembly from an individual Lepidonotus clava (scale worm; Annelida; Polychaeta; Phyllodocida; Polynoidae). The genome sequence is 1,044 megabases in span. Most of the assembly is scaffolded into 18 chromosomal pseudomolecules. The mitochondrial genome has also been assembled and is 15.6 kilobases in length.
Keywords: Lepidonotus clava; Polychaeta; chromosomal; genome sequence; scale worm.
Copyright: © 2022 Darbyshire T et al.
Conflict of interest statement
No competing interests were disclosed.
Figures
Similar articles
-
The genome sequence of the Gelatinous Scale Worm, Alentia gelatinosa (Sars, 1835).Wellcome Open Res. 2023 Nov 22;8:542. doi: 10.12688/wellcomeopenres.20176.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 38558923 Free PMC article.
-
The genome sequence of a scale worm, Harmothoe impar (Johnston, 1839).Wellcome Open Res. 2023 Jul 20;8:315. doi: 10.12688/wellcomeopenres.19570.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 37786780 Free PMC article.
-
The genome sequence of the segmented worm, Sthenelais limicola (Ehlers, 1864).Wellcome Open Res. 2023 Jan 19;8:31. doi: 10.12688/wellcomeopenres.18856.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 37469856 Free PMC article.
-
The genome sequence of the King Ragworm, Alitta virens (Sars, 1835).Wellcome Open Res. 2023 Jul 11;8:297. doi: 10.12688/wellcomeopenres.19642.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 37469858 Free PMC article.
-
The genome sequence of a segmented worm, Terebella lapidaria Linnaeus, 1767.Wellcome Open Res. 2024 Aug 7;9:432. doi: 10.12688/wellcomeopenres.22823.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 39221441 Free PMC article.
Cited by
-
Annelid Comparative Genomics and the Evolution of Massive Lineage-Specific Genome Rearrangement in Bilaterians.Mol Biol Evol. 2024 Sep 4;41(9):msae172. doi: 10.1093/molbev/msae172. Mol Biol Evol. 2024. PMID: 39141777 Free PMC article.
-
Genomic Analysis of a Scale Worm Provides Insights into Its Adaptation to Deep-Sea Hydrothermal Vents.Genome Biol Evol. 2023 Jul 3;15(7):evad125. doi: 10.1093/gbe/evad125. Genome Biol Evol. 2023. PMID: 37401460 Free PMC article.
-
The genome sequence of the Gelatinous Scale Worm, Alentia gelatinosa (Sars, 1835).Wellcome Open Res. 2023 Nov 22;8:542. doi: 10.12688/wellcomeopenres.20176.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 38558923 Free PMC article.
-
Acceleration of genome rearrangement in clitellate annelids.bioRxiv [Preprint]. 2024 May 14:2024.05.12.593736. doi: 10.1101/2024.05.12.593736. bioRxiv. 2024. PMID: 38798472 Free PMC article. Preprint.
References
-
- Barnich R, Fiege D: The Aphroditoidea (Annelida: Polychaeta) of the Mediterranean Sea. Abhandlungen der Senckenbergischen Naturforschenden Gesellschaft. 2003;1–167. Reference Source
-
- Chambers SJ, Muir AI: Polychaetes: British Chrysopetaloidea, Pisionoidea and Aphroditoidea. Synopses of the British Fauna (New Series). 1997;54:1–202. Reference Source
Grants and funding
LinkOut - more resources
Full Text Sources

