Antisense oligonucleotide rescue of CGG expansion-dependent FMR1 mis-splicing in fragile X syndrome restores FMRP
- PMID: 37364131
- PMCID: PMC10319035
- DOI: 10.1073/pnas.2302534120
Antisense oligonucleotide rescue of CGG expansion-dependent FMR1 mis-splicing in fragile X syndrome restores FMRP
Abstract
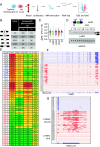
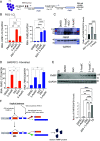
Aberrant alternative splicing of mRNAs results in dysregulated gene expression in multiple neurological disorders. Here, we show that hundreds of mRNAs are incorrectly expressed and spliced in white blood cells and brain tissues of individuals with fragile X syndrome (FXS). Surprisingly, the FMR1 (Fragile X Messenger Ribonucleoprotein 1) gene is transcribed in >70% of the FXS tissues. In all FMR1-expressing FXS tissues, FMR1 RNA itself is mis-spliced in a CGG expansion-dependent manner to generate the little-known FMR1-217 RNA isoform, which is comprised of FMR1 exon 1 and a pseudo-exon in intron 1. FMR1-217 is also expressed in FXS premutation carrier-derived skin fibroblasts and brain tissues. We show that in cells aberrantly expressing mis-spliced FMR1, antisense oligonucleotide (ASO) treatment reduces FMR1-217, rescues full-length FMR1 RNA, and restores FMRP (Fragile X Messenger RibonucleoProtein) to normal levels. Notably, FMR1 gene reactivation in transcriptionally silent FXS cells using 5-aza-2'-deoxycytidine (5-AzadC), which prevents DNA methylation, increases FMR1-217 RNA levels but not FMRP. ASO treatment of cells prior to 5-AzadC application rescues full-length FMR1 expression and restores FMRP. These findings indicate that misregulated RNA-processing events in blood could serve as potent biomarkers for FXS and that in those individuals expressing FMR1-217, ASO treatment may offer a therapeutic approach to mitigate the disorder.
Keywords: FMR1; FMRP; RNA splicing; antisense oligonucleotides.
Conflict of interest statement
S.S., E.B.-K., and J.D.R., have filed a patent on RNA-based biomarkers for Fragile X associated disorders. S.S., J.K.W., and J.D.R. have filed a patent on ASO based therapeutics for Fragile X associated disorders.
Figures



References
-
- Darnell J. C., et al. , FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 14, 247–261 (2011), https://doi.org/10.1016/j.cell.2011.06.013. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

