Border-associated macrophages mediate the neuroinflammatory response in an alpha-synuclein model of Parkinson disease
- PMID: 37365181
- PMCID: PMC10293214
- DOI: 10.1038/s41467-023-39060-w
Border-associated macrophages mediate the neuroinflammatory response in an alpha-synuclein model of Parkinson disease
Abstract
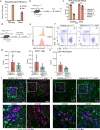
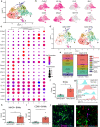
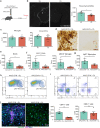
Dopaminergic cell loss due to the accumulation of α-syn is a core feature of the pathogenesis of Parkinson disease. Neuroinflammation specifically induced by α-synuclein has been shown to exacerbate neurodegeneration, yet the role of central nervous system (CNS) resident macrophages in this process remains unclear. We found that a specific subset of CNS resident macrophages, border-associated macrophages (BAMs), play an essential role in mediating α-synuclein related neuroinflammation due to their unique role as the antigen presenting cells necessary to initiate a CD4 T cell response whereas the loss of MHCII antigen presentation on microglia had no effect on neuroinflammation. Furthermore, α-synuclein expression led to an expansion in border-associated macrophage numbers and a unique damage-associated activation state. Through a combinatorial approach of single-cell RNA sequencing and depletion experiments, we found that border-associated macrophages played an essential role in immune cell recruitment, infiltration, and antigen presentation. Furthermore, border-associated macrophages were identified in post-mortem PD brain in close proximity to T cells. These results point to a role for border-associated macrophages in mediating the pathogenesis of Parkinson disease through their role in the orchestration of the α-synuclein-mediated neuroinflammatory response.
© 2023. The Author(s).
Conflict of interest statement
Dr. Standaert has served as a consultant for or received honoraria from Abbvie Inc., Curium Pharma, Appello, Theravance, Sanofi-Aventis, Alnylam Pharmaceutics, Coave Therapeutics, BlueRock Therapeutics, and F. Hoffman-La Roche. All remaining authors declare no competing interests.
Figures



Comment in
-
CNS-associated macrophages shape the inflammatory response in a mouse model of Parkinson's disease.Nat Commun. 2023 Jun 26;14(1):3753. doi: 10.1038/s41467-023-39061-9. Nat Commun. 2023. PMID: 37365158 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

