Interferon signaling promotes tolerance to chromosomal instability during metastatic evolution in renal cancer
- PMID: 37365326
- PMCID: PMC10368532
- DOI: 10.1038/s43018-023-00584-1
Interferon signaling promotes tolerance to chromosomal instability during metastatic evolution in renal cancer
Abstract
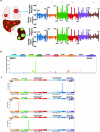
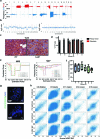
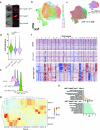
Molecular routes to metastatic dissemination are critical determinants of aggressive cancers. Through in vivo CRISPR-Cas9 genome editing, we generated somatic mosaic genetically engineered models that faithfully recapitulate metastatic renal tumors. Disruption of 9p21 locus is an evolutionary driver to systemic disease through the rapid acquisition of complex karyotypes in cancer cells. Cross-species analysis revealed that recurrent patterns of copy number variations, including 21q loss and dysregulation of the interferon pathway, are major drivers of metastatic potential. In vitro and in vivo genomic engineering, leveraging loss-of-function studies, along with a model of partial trisomy of chromosome 21q, demonstrated a dosage-dependent effect of the interferon receptor genes cluster as an adaptive mechanism to deleterious chromosomal instability in metastatic progression. This work provides critical knowledge on drivers of renal cell carcinoma progression and defines the primary role of interferon signaling in constraining the propagation of aneuploid clones in cancer evolution.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures













Comment in
-
Interferon signaling restrains renal cell carcinoma heterogeneity.Trends Cancer. 2023 Nov;9(11):871-873. doi: 10.1016/j.trecan.2023.08.008. Epub 2023 Aug 30. Trends Cancer. 2023. PMID: 37658022 Free PMC article.
References
-
- Dabestani S, et al. Renal cell carcinoma recurrences and metastases in primary non-metastatic patients: a population-based study. World J. Urol. 2016;34:1081–1086. - PubMed
-
- Kotecha RR, Motzer RJ, Voss MH. Towards individualized therapy for metastatic renal cell carcinoma. Nat. Rev. Clin. Oncol. 2019;16:621–633. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

