The microbiota of pregnant women with SARS-CoV-2 and their infants
- PMID: 37365606
- PMCID: PMC10291758
- DOI: 10.1186/s40168-023-01577-z
The microbiota of pregnant women with SARS-CoV-2 and their infants
Abstract
Background: Infants receive their first bacteria from their birthing parent. This newly acquired microbiome plays a pivotal role in developing a robust immune system, the cornerstone of long-term health.
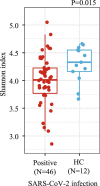
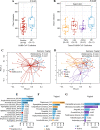
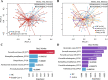
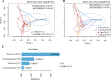
Results: We demonstrated that the gut, vaginal, and oral microbial diversity of pregnant women with SARS-CoV-2 infection is reduced, and women with early infections exhibit a different vaginal microbiota composition at the time of delivery compared to their healthy control counterparts. Accordingly, a low relative abundance of two Streptococcus sequence variants (SV) was predictive of infants born to pregnant women with SARS-CoV-2 infection.
Conclusions: Our data suggest that SARS-CoV-2 infections during pregnancy, particularly early infections, are associated with lasting changes in the microbiome of pregnant women, compromising the initial microbial seed of their infant. Our results highlight the importance of further exploring the impact of SARS-CoV-2 on the infant's microbiome-dependent immune programming. Video Abstract.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

