Transcriptionally regulated miR-26a-5p may act as BRCAness in Triple-Negative Breast Cancer
- PMID: 37365643
- PMCID: PMC10294332
- DOI: 10.1186/s13058-023-01663-y
Transcriptionally regulated miR-26a-5p may act as BRCAness in Triple-Negative Breast Cancer
Abstract
Background: DNA damage and DNA damage repair (DDR) are important therapeutic targets for triple-negative breast cancer (TNBC), a subtype with limited chemotherapy efficiency and poor outcome. However, the role of microRNAs in the therapy is emerging. In this study, we explored whether miR-26a-5p could act as BRCAness and enhance chemotherapy sensitivity in TNBC.
Methods: Quantitative reverse transcription polymerase chain reaction (RT-qPCR) was used to detect the expression of miR-26a-5p in breast cancer tissues and cell lines. CCK-8 was used to measure drug sensitivity in concentration gradient and time gradient. Comet assay was used to detect DNA damage. Flow cytometry was performed to examine apoptosis. Moreover, we used western blot and immunofluorescence to detect biomarkers. Luciferase reporter assay was performed to verify the combination of miR-26a-5p and 3'UTR of target gene. Hormone deprivation and stimulation assay were used to validate the effect of hormone receptors on the expression of miR-26a-5p. Chromatin immunoprecipitation (ChIP) assays were used to verify the binding sites of ER-a or PR with the promoter of miR-26a-5p. Animal experiments were performed to the effect of miR-26a-5p on Cisplatin treatment.
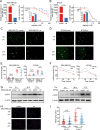
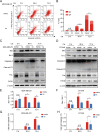
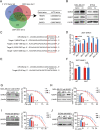
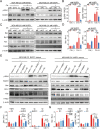
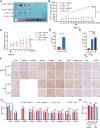
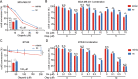
Results: The expression of miR-26a-5p was significantly downregulated in TNBC. Overexpressing miR-26a-5p enhanced the Cisplatin-induced DNA damage and following apoptosis. Interestingly, miR-26a-5p promoted the expression of Fas without Cisplatin stimulating. It suggested that miR-26a-5p provided a hypersensitivity state of death receptor apoptosis and promoted the Cisplatin sensitivity of TNBC cells in vitro and in vivo. Besides, miR-26a-5p negatively regulated the expression of BARD1 and NABP1 and resulted in homologous recombination repair defect (HRD). Notably, overexpressing miR-26a-5p not only facilitated the Olaparib sensitivity of TNBC cells but also the combination of Cisplatin and Olaparib. Furthermore, hormone receptors functioned as transcription factors in the expression of miR-26a-5p, which explained the reasons that miR-26a-5p expressed lowest in TNBC.
Conclusions: Taken together, we reveal the important role of miR-26a-5p in Cisplatin sensitivity and highlight its new mechanism in DNA damage and synthetic lethal.
Keywords: Cisplatin; DNA damage; HRD; Synthetic lethal; TNBC; miR-26a-5p.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

