Plasma fractalkine contributes to systemic myeloid diversity and PD-L1/PD-1 blockade in lung cancer
- PMID: 37366231
- PMCID: PMC10398648
- DOI: 10.15252/embr.202255884
Plasma fractalkine contributes to systemic myeloid diversity and PD-L1/PD-1 blockade in lung cancer
Abstract
Recent studies highlight the importance of baseline functional immunity for immune checkpoint blockade therapies. High-dimensional systemic immune profiling is performed in a cohort of non-small-cell lung cancer patients undergoing PD-L1/PD-1 blockade immunotherapy. Responders show high baseline myeloid phenotypic diversity in peripheral blood. To quantify it, we define a diversity index as a potential biomarker of response. This parameter correlates with elevated activated monocytic cells and decreased granulocytic phenotypes. High-throughput profiling of soluble factors in plasma identifies fractalkine (FKN), a chemokine involved in immune chemotaxis and adhesion, as a biomarker of response to immunotherapy that also correlates with myeloid cell diversity in human patients and murine models. Secreted FKN inhibits lung adenocarcinoma growth in vivo through a prominent contribution of systemic effector NK cells and increased tumor immune infiltration. FKN sensitizes murine lung cancer models refractory to anti-PD-1 treatment to immune checkpoint blockade immunotherapy. Importantly, recombinant FKN and tumor-expressed FKN are efficacious in delaying tumor growth in vivo locally and systemically, indicating a potential therapeutic use of FKN in combination with immunotherapy.
Keywords: NK cells; adenocarcinoma; biomarker; monocytes; neutrophils.
© 2023 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

- A
Diversity indexes as a function of clinical responses. Each dot represents a biological replicate.
- B
ROC curve of the diversity index as a function of objective responses vs disease progression.
- C
Kaplan–Meier plot of PFS stratifying the patients according to high or low diversity index. N = 69 (33 above the cut‐off, 36 below).
- D
As in (C) but plotting OS. N = 72 (34 above the cut‐off, 38 below).
- E
Correlation between the percentage of monocytes and diversity index. Each dot represents a biological replicate.
- F
Correlation between the percentage of granulocytic myeloid cells and diversity index. Each dot represents a biological replicate.
- G
Baseline plasma FKN concentrations in responders (R, n = 26), progressors (PR, n = 62), hyperprogressors (HPR, n = 3), age‐matched healthy donors (H, n = 32), and patients who were not eligible for immunotherapy (NO IT, n = 28). Sample sizes represent biological replicates. Error bars are shown (standard deviations, SD), and relevant statistical comparisons by Mann–Whitney U tests are shown in the graphs.
- H
Correlation between FKN plasma concentration and diversity index with the Spearman's test. Each dot represents a biological replicate.
- I
ROC analysis of FKN concentration as a predictor of objective clinical responses. The calculated cut‐off value for the statistics provided in the graph is shown.
- J
Kaplan–Meier plot of PFS stratifying the patients according to the FKN cut‐off value identified in the ROC curve (n = 42). The associated P‐value is shown.
- K
As in (J) but plotting OS. n = 46 (17 above the cut‐off, 29 below).
- L
Time elapsed from disease diagnosis to the beginning of immunotherapy in second‐line treated patients. Each dot represents a biological replicate.

- A–D
Representative SPADE3 cluster profiles of myeloid cells integrating 43 markers are shown for (A) a long‐term responder, (B) a short‐term responder (stable disease), (C) a progressor (PR), and (D) a hyperprogressor (HPR). Distributions of CD14 (left dendrogram) and CD66b expression (right dendrogram) are shown. Main cell subsets are encircled and identified as Mo—monocytes; Neu—neutrophils; G‐MDSC—granulocytic myeloid‐derived suppressor cells; NC‐Mo—nonclassical monocytes; NK—natural killer cells. The relative expression of the selected marker as indicated above the graphs color‐coded, from dark red (maximum expression) to dark blue (minimum expression).

- A
Left graph, baseline frequency of monocytes (CD14+ HLA‐DR+) vs neutrophils (CD14+ CD66b+) within CD11b+ cells in patients classified as responders (R, green), progressors (PR, red), stable disease (blue) and hyperprogressors (purple). The number of responders and progressors is indicated in each quadrant. Center graph, as in left but plotting the percentage of G‐MDSCs (CD14− CD66b+) vs monocytes. Right graph, as in left but plotting the percentage of Mo‐MDSC (CD14+ HLA‐DR−) vs. monocytes.
- B
Left graph, relative percentages of the main myeloid populations restricted to this compartment are plotted for each patient under study as indicated as color codes, classified according to objective responders and stable disease (SD); Right graph, as in left but in progressors and hyperprogressors (HPR).
- C
Percentage of circulating monocytes (CD11b+ CD14+ HLA‐DR+) within each response group as indicated.
- D
Percentage of granulocytic cells (CD11b+ CD66b+) within each response group as indicated.
- E
ROC analysis of the percentage of monocytes as a predictor of objective responses.
- F
ROC analysis of the percentage of granulocytic myeloid cells as a predictor of no objective response.

- A
Baseline plasma FKN concentrations in responders (R, n = 13) and progressors (PR, n = 5) in a validation cohort of NSCLC patients treated with anti‐PD‐1/PD‐L1 immunotherapy as a first‐line treatment. Error bars are shown (standard deviations, SD). Statistical significance was tested by the chi‐square test.
- B, C
(B) Representative double lung immunostainings for Pan‐Cytokeratin (PanCK) (brown) and FKN (blue; red arrows indicate FKN positive cells) in lung tissue sections from LUAD patients and (C) adjacent healthy tissue.
- D, E
(D) Pearson's correlation of PanCK with FKN positive areas (%) in histologies of adjacent nontumor and (E) LUAD tissues. Pearson's correlation coefficients are shown in the graphs. n = 10.
- F
FKN mRNA expression level in tumor (T) and normal tissue (N) of clinical samples from lung adenocarcinoma (LUAD) and squamous lung carcinoma (LUSC) patients registered in the TCGA database. Distributions of gene expression levels are displayed using box plots. Box and whisker plots indicate median (central line), 25th to 75th percentiles (box), and minimum to maximum values (whiskers).
- G
Top, lentivectors for the expression of cytokines of interest. SIN, self‐inactivating deleted LTR; LTR, long‐terminal repeat; SFFVp, spleen focus‐forming virus promoter; UBIp, human ubiquitin promoter; Puro R, puromycin resistance gene. Down, ELISA quantification of cytokine secretion by the indicated engineered lung cancer cell lines expressing the indicated cytokines (T). Endogenous secretion of each cytokine was also quantified in supernatants from cultures of parental unmodified controls (UT). Data are presented as mean ± SD (n = 3 independent biological replicates).
- H
Real‐time cell growth (RTCA) of 3LL cell lines engineered to secrete the indicated cytokines. Relevant statistical comparisons of delta‐cell indexes after 50 h of culture were carried out by ANOVA. Data are presented as mean ± SD (n = 3 independent biological replicates).
- I
Real‐time cell growth (RTCA) of unmodified and FKN‐producing 3LL cell lines cultured with 1 μg/ml of recombinant FKN (rFKN).
- J
Flow cytometric assessment of propidium iodide (PI) uptake by apoptotic unmodified 3LL cells cultured in 3LL‐WT (control) or 3LL‐FKN (FKN) conditioned medium for 24, 48, or 72 h.
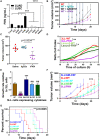
- A
Secreted FKN in cell cultures of a collection of 10 LUAD and LUSC cancer cell lines quantified by ELISA. Results are presented as mean ± SD. Samples were assayed in duplicates from a pool of three independent replicates. Statistical differences among groups were analyzed by ANOVA followed by Tukey's tests.
- B
Tumor growth curves of 3LL tumor‐bearing mice treated with recombinant FKN (rFKN), the CX3CR1 inhibitor AZD8797, a nonrelevant protein (IgG2a) or saline buffer as vehicle control (WT). Arrows indicate the time of intraperitoneal injections of rFKN (blue) and AZD8797 (black). Data are expressed as mean ± SD (n = 6 mice per group). Statistical comparisons by ANOVA followed by Tukey's pairwise comparison tests are provided.
- C
Dot plot of 3LL tumor size in the indicated groups of mice at day 10 after cancer cell inoculation. Statistical comparisons by ANOVA and Tukey's pairwise comparison tests are indicated.
- D
Proliferation of two LUAD murine cell lines overexpressing FKN and their parental unmodified counterparts (WT), as indicated. Real‐time cell growth was monitored by RTCA. Delta‐cell indexes are expressed as mean ± SD from three independent replicates. Differences in delta‐cell index proliferation data were tested by ANOVA following Tukey's pairwise comparison tests after 40 h of culture.
- E
Bar graphs with cell growth rates for the indicated 3LL cell lines relative to the growth of unmodified 3LL cells. Error bars are shown (SD). Statistical comparisons among three independent biological replicates by ANOVA and Tukey's pairwise comparison tests are indicated.
- F
In vivo tumor growth of engrafted 3LL cell lines producing the indicated cytokines. Data are expressed as mean ± SD (n = 6 mice per group). Comparisons between groups were performed by ANOVA and Tukey's pairwise comparison tests.
- G
Kaplan–Meier survival plots. Differences between the control group (WT) and the FKN group were evaluated by a two‐sided log‐rank test.

- A
FKN plasma concentration at day 15 in the indicated groups of tumor‐bearing mice. Data are expressed as mean ± SD from a pool of 6 mice/group. Statistical comparisons were performed by ANOVA and Tukey's pairwise comparison tests.
- B
Tumor volumes 15 days after tumor inoculation. Data are expressed as mean ± SD (n = 6 mice per group), and comparisons between groups were performed by ANOVA and Tukey's pairwise comparison tests.
- C
Relative myeloid and lymphoid composition of peripheral blood at day 15 in mice transplanted with 3LL‐expressing the indicated cytokines. WT, unmodified 3LL cells; Healthy, mice without tumors.
- D
Percentage of the indicated peripheral immune cell populations at day 15 after injection of groups of mice (n = 6 mice per group) with 3LL cells overexpressing the indicated myeloid‐regulating cytokines. The relevant immune populations were quantified as percentages of total leukocytes (CD45+ cells), specifying DCs (CD11c), macrophages (F4/80), Ly6C+ monocytes, and Ly6C− monocytes, granulocytes (Ly6G), B cells (CD19), NKs (NK1.1), CD4 T cells (CD3 CD4) and CD8 T cells (CD3 CD8). Relevant statistical comparisons were performed by the Wilcoxon's test followed by pairwise comparisons of relevance by the Mann–Whitney U test. Box and whisker plots indicate median (central line), 25th to 75th percentiles (box), and minimum to maximum values (whiskers).

- A
Dot plot of tumor volume in mice 22 days after engraftment of the indicated cancer cell lines and subjected to either a control treatment or anti‐PD‐1 therapy. Relevant statistical comparisons are shown in the graph by the U of Mann–Whitney test.
- B
Unmodified and FKN‐expressing 3LL cells were subcutaneously inoculated in mice and tumors were allowed to grow for 7 days. Tumor‐engrafted mice were treated intraperitoneally with anti‐PD‐1 antibody or vehicle (days 7, 11, and 15, as indicated by arrows) following randomization into two groups. Tumor growth was monitored. Data are presented as mean ± SD (n = 6 mice per group).
- C
Kaplan–Meier survival plots of the indicated groups of mice. Survival differences between the control group (WT + αPD‐1) and FKN + αPD‐1 were evaluated by a two‐sided log‐rank test.
- D–H
(D) Percentage of splenic Ly6C+ monocytes, (E) Ly6G− monocytes, (F) Ly6G+ neutrophils, (G) PD‐1+ T cells (CD3+), and (H) PD‐1+ NK cells (NK1.1+). Data are presented as mean ± SD (n = 6 mice per group).
- I–K
(I) Percentage of tumor infiltration with total leukocytes (CD45+), (J) CD4 T cells, and (K) NK cells (NK1.1+). Infiltration data are shown as mean ± SD (n = 8 mice per group).

- A
The graphs represent percentages of the indicated infiltrating immune cell types as quantified by flow cytometry, in spleens obtained from mice inoculated with the indicated cell lines (parental cell line, WT; 3LL cells expressing FKN, FKN) with or without PD‐1 blockade treatment. CD4 and CD8 T lymphocytes, NK cells (NK1.1), B cells (CD19), MDSCs (Ly6G+ CD115+), macrophages (F4/80), and DCs (CD11c) were quantified at day 14 after tumor inoculation. Data are shown as the mean of the percentage within total leukocytes (CD45+) ± SD (n = 6 mice). Relevant statistical comparisons are shown in the graphs, evaluated by ANOVA and Tukey's pairwise comparisons. *, ***, indicate significant (P < 0.05) and highly significant (P < 0.001) differences. ns, nonsignificant differences.
- B
Graphs represent percentages of the indicated infiltrating immune cell types as quantified by flow cytometry, in tumors excised from mice inoculated with the indicated cell lines (parental cell line, WT; 3LL cells expressing FKN, FKN) with or without PD‐1 blockade. Neutrophils (Ly6G), CD8, B cells (CD19), Ly6C+ monocytes, DCs (CD11c), and macrophages (F4/80) were quantified at day 14 after tumor inoculation. Data are shown as the mean of the percentage within total leukocytes (CD45+) ± SD (n = 8 mice). Relevant statistical comparisons are shown in the graphs, evaluated by ANOVA and Tukey's pairwise comparisons. ns, nonsignificant differences.

- A
Evaluation of tumor infiltration with the indicated selected immune cell populations, and correlation analyses of FKN transcriptional expression and immune infiltrates from the TCGA database. Analyses were restricted to lung adenocarcinoma samples (n = 515). Spearman correlation with different immune populations was identified by several algorithms (CIBERSORT, quanTIseq, xCell, TIDE) and an adjustment based on tumor purity was employed to minimize the potential interaction of low tumor cell quantities. Each dot represents a single tumor sample. Spearman's rho value and P‐values are provided within the graphs.
- B
Correlation between tumor FKN and PD‐L1 transcriptional expression by the Spearman's test. Relevant statistical results are presented within the graphs.
- C
As in (B) but plotting PD‐1 transcriptional expression levels.

- A
Experimental schedule for testing systemic FKN anti‐tumor activities. 3LL‐parental (WT) or 3LL‐FKN (FKN) cells were subcutaneously injected into the left flank of mice (inoculation side). Seven days later, 3LL‐parental cells were engrafted on the right flank (target tumor side). Mice were intraperitoneally treated with anti‐PD‐1 antibody at days 7, 11, and 15 after the last tumor inoculation.
- B
Tumor growth of right and left flank engrafted tumors in the experimental schedule shown in (A). Data are presented as mean ± SD (n = 6 mice per group).
- C
Tumor growth in mice intraperitoneally treated with anti‐CD4, anti‐CD8, and anti‐NK1.1 depleting antibodies at days 6, 10, 14, and 18 after tumor inoculation, as indicated by arrows. Data are presented as mean ± SD (n = 6 mice per group).
- D
Kaplan–Meier survival plot of the indicated treatment groups. Survival differences between the control group (WT) and FKN were evaluated by a two‐sided log‐rank test.
- E–J
(E) Percentage of splenic NKs (NK1.1+) expressing KLRG1, (F) TIGIT, and (G) TIM3. (H) Percentage of splenic T cells (CD3+) expressing KLRG1, (I) TIGIT, and (J) TIM3. Data are presented as mean ± SD (n = 6 mice per group). Comparisons among groups were performed by ANOVA and Tukey's pairwise comparison tests.
References
-
- Ajona D, Ortiz‐Espinosa S, Lozano T, Exposito F, Calvo A, Valencia K, Redrado M, Remírez A, Lecanda F, Alignani D et al (2020) Short‐term starvation reduces IGF‐1 levels to sensitize lung tumors to PD‐1 immune checkpoint blockade. Nat Cancer 1: 75–85 - PubMed
-
- Arasanz H, Bocanegra A, Morilla I, Fernandez‐Irigoyen J, Martinez‐Aguillo M, Teijeira L, Garnica M, Blanco E, Chocarro L, Ausin K et al (2022) Circulating low density neutrophils are associated with resistance to first‐line anti‐PD1/PDL1 immunotherapy in non‐small cell lung cancer. Cancers (Basel) 14: 3846 - PMC - PubMed
-
- Bleau AM, Freire J, Pajares MJ, Zudaire I, Anton I, Nistal‐Villan E, Redrado M, Zandueta CN, Garmendia I, Ajona D et al (2014) New syngeneic inflammatory‐related lung cancer metastatic model harboring double KRAS/WWOX alterations. Int J Cancer 135: 2516–2527 - PubMed
-
- Bocanegra A, Fernandez‐Hinojal G, Zuazo‐Ibarra M, Arasanz H, Garcia‐Granda MJ, Hernandez C, Ibanez M, Hernandez‐Marin B, Martinez‐Aguillo M, Lecumberri MJ et al (2019) PD‐L1 expression in systemic immune cell populations as a potential predictive biomarker of responses to PD‐L1/PD‐1 blockade therapy in lung cancer. Int J Mol Sci 20: 1631 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

