Nonsense-mediated decay machinery in Plasmodium falciparum is inefficient and non-essential
- PMID: 37366629
- PMCID: PMC10449492
- DOI: 10.1128/msphere.00233-23
Nonsense-mediated decay machinery in Plasmodium falciparum is inefficient and non-essential
Abstract
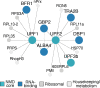
Nonsense-mediated decay (NMD) is a conserved mRNA quality control process that eliminates transcripts bearing a premature termination codon. In addition to its role in removing erroneous transcripts, NMD is involved in post-transcriptional regulation of gene expression via programmed intron retention in metazoans. The apicomplexan parasite Plasmodium falciparum shows relatively high levels of intron retention, but it is unclear whether these variant transcripts are functional targets of NMD. In this study, we use CRISPR-Cas9 to disrupt and epitope-tag the P. falciparum orthologs of two core NMD components: PfUPF1 (PF3D7_1005500) and PfUPF2 (PF3D7_0925800). We localize both PfUPF1 and PfUPF2 to puncta within the parasite cytoplasm and show that these proteins interact with each other and other mRNA-binding proteins. Using RNA-seq, we find that although these core NMD orthologs are expressed and interact in P. falciparum, they are not required for degradation of nonsense transcripts. Furthermore, our work suggests that the majority of intron retention in P. falciparum has no functional role and that NMD is not required for parasite growth ex vivo. IMPORTANCE In many organisms, the process of destroying nonsense transcripts is dependent on a small set of highly conserved proteins. We show that in the malaria parasite, these proteins do not impact the abundance of nonsense transcripts. Furthermore, we demonstrate efficient CRISPR-Cas9 editing of the malaria parasite using commercial Cas9 nuclease and synthetic guide RNA, streamlining genomic modifications in this genetically intractable organism.
Keywords: RNA-seq; intron retention; mRNA degradation; malaria; nonsense-mediated decay.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

