Imunocapture Magnetic Beads Enhanced and Ultrasensitive CRISPR-Cas13a-Assisted Electrochemical Biosensor for Rapid Detection of SARS-CoV-2
- PMID: 37366962
- PMCID: PMC10296353
- DOI: 10.3390/bios13060597
Imunocapture Magnetic Beads Enhanced and Ultrasensitive CRISPR-Cas13a-Assisted Electrochemical Biosensor for Rapid Detection of SARS-CoV-2
Abstract
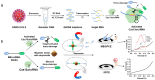
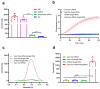
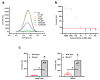
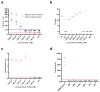
The rapid and ongoing spread of the coronavirus disease (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emphasizes the urgent need for an easy and sensitive virus detection method. Here, we describe an immunocapture magnetic bead-enhanced electrochemical biosensor for ultrasensitive SARS-CoV-2 detection based on clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins, collectively known as CRISPR-Cas13a technology. At the core of the detection process, low-cast and immobilization-free commercial screen-printed carbon electrodes are used to measure the electrochemical signal, while streptavidin-coated immunocapture magnetic beads are used to reduce the background noise signal and enhance detection ability by separating the excessive report RNA, and a combination of isothermal amplification methods in the CRISPR-Cas13a system is used for nucleic acid detection. The results showed that the sensitivity of the biosensor increased by two orders of magnitude when the magnetic beads were used. The proposed biosensor required approximately 1 h of overall processing time and demonstrated an ultrasensitive ability to detect SARS-CoV-2, which could be as low as 1.66 aM. Furthermore, owing to the programmability of the CRISPR-Cas13a system, the biosensor can be flexibly applied to other viruses, providing a new approach for powerful clinical diagnostics.
Keywords: CRISPR Cas13a; bioanalytical chemistry; biosensor; electrochemistry; trans-acting cleavage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Electrochemical biosensor for nucleic acid amplification-free and sensitive detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA via CRISPR/Cas13a trans-cleavage reaction.Biosens Bioelectron. 2022 Apr 1;201:113960. doi: 10.1016/j.bios.2021.113960. Epub 2022 Jan 4. Biosens Bioelectron. 2022. PMID: 35016109 Free PMC article.
-
Ultrasensitive and amplification-free detection of SARS-CoV-2 RNA using an electrochemical biosensor powered by CRISPR/Cas13a.Bioelectrochemistry. 2023 Apr;150:108364. doi: 10.1016/j.bioelechem.2023.108364. Epub 2023 Jan 4. Bioelectrochemistry. 2023. PMID: 36621051 Free PMC article.
-
Amplification-Free Detection of SARS-CoV-2 and Respiratory Syncytial Virus Using CRISPR Cas13a and Graphene Field-Effect Transistors.Angew Chem Int Ed Engl. 2022 Aug 8;61(32):e202203826. doi: 10.1002/anie.202203826. Epub 2022 May 31. Angew Chem Int Ed Engl. 2022. PMID: 35559592 Free PMC article.
-
[Recent advances in clustered regularly interspaced short palindromic repeats-based detection of severe acute respiratory syndrome coronavirus 2].Se Pu. 2022 Sep;40(9):773-781. doi: 10.3724/SP.J.1123.2022.08001. Se Pu. 2022. PMID: 36156623 Free PMC article. Review. Chinese.
-
CRISPR-Cas based virus detection: Recent advances and perspectives.Biosens Bioelectron. 2021 Dec 1;193:113541. doi: 10.1016/j.bios.2021.113541. Epub 2021 Aug 8. Biosens Bioelectron. 2021. PMID: 34418634 Free PMC article. Review.
Cited by
-
Current Trends in RNA Virus Detection via Nucleic Acid Isothermal Amplification-Based Platforms.Biosensors (Basel). 2024 Feb 11;14(2):97. doi: 10.3390/bios14020097. Biosensors (Basel). 2024. PMID: 38392016 Free PMC article. Review.
-
Mitigating Antibiotic Resistance: The Utilization of CRISPR Technology in Detection.Biosensors (Basel). 2024 Dec 20;14(12):633. doi: 10.3390/bios14120633. Biosensors (Basel). 2024. PMID: 39727898 Free PMC article. Review.
-
CRISPR-Based Biosensors for Medical Diagnosis: Readout from Detector-Dependence Detection Toward Naked Eye Detection.Biosensors (Basel). 2024 Jul 28;14(8):367. doi: 10.3390/bios14080367. Biosensors (Basel). 2024. PMID: 39194596 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

