A Decade-Long Evaluation of Neonatal Septicaemic Escherichia coli: Clonal Lineages, Genomes, and New Delhi Metallo-Beta-Lactamase Variants
- PMID: 37367488
- PMCID: PMC10434172
- DOI: 10.1128/spectrum.05215-22
A Decade-Long Evaluation of Neonatal Septicaemic Escherichia coli: Clonal Lineages, Genomes, and New Delhi Metallo-Beta-Lactamase Variants
Abstract
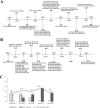
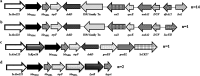
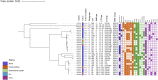
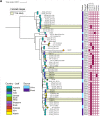
Longitudinal studies of extraintestinal pathogenic Escherichia coli (ExPEC) and epidemic clones of E. coli in association with New Delhi metallo-β-lactamase (blaNDM) in septicaemic neonates are rare. This study captured the diversity of 80 E. coli isolates collected from septicaemic neonates in terms of antibiotic susceptibility, resistome, phylogroups, sequence types (ST), virulome, plasmids, and integron types over a decade (2009 to 2019). Most of the isolates were multidrug-resistant and, 44% of them were carbapenem-resistant, primarily due to blaNDM. NDM-1 was the sole NDM-variant present in conjugative IncFIA/FIB/FII replicons until 2013, and it was subsequently replaced by other variants, such as NDM-5/-7 found in IncX3/FII. A core genome analysis for blaNDM+ve isolates showed the heterogeneity of the isolates. Fifty percent of the infections were caused by isolates of phylogroups B2 (34%), D (11.25%), and F (4%), whereas the other half were caused by phylogroups A (25%), B1 (11.25%), and C (14%). The isolates were further distributed in approximately 20 clonal complexes (STC), including five epidemic clones (ST131, ST167, ST410, ST648, and ST405). ST167 and ST131 (subclade H30Rx) were dominant, with most of the ST167 being blaNDM+ve and blaCTX-M-15+ve. In contrast, the majority of ST131 isolates were blaNDM-ve but blaCTX-M-15+ve, and they possessed more virulence determinants than did ST167. A single nucleotide polymorphism (SNP)-based comparative genome analysis of epidemic clones ST167 and ST131 in a global context revealed that the study isolates were present in close proximity but were distant from global isolates. The presence of antibiotic-resistant epidemic clones causing sepsis calls for a modification of the recommended antibiotics with which to treat neonatal sepsis. IMPORTANCE Multidrug-resistant and virulent ExPEC causing sepsis in neonates is a challenge to neonatal health. The presence of enzymes, such as carbapenemases (blaNDM) that hydrolyze most β-lactam antibiotic compounds, result in difficulties when treating neonates. The characterization of ExPECs collected over 10 years showed that 44% of ExPECs were carbapenem-resistant, possessing transmissible blaNDM genes. The isolates belonged to different phylogroups that are considered to be either commensals or virulent. The isolates were distributed in around 20 clonal complexes (STC), including two predominant epidemic clones (ST131 and ST167). ST167 possessed few virulence determinants but was blaNDM+ve. In contrast, ST131 harbored several virulence determinants but was blaNDM-ve. A comparison of the genomes of these epidemic clones in a global context revealed that the study isolates were present in close proximity but were distant from global isolates. The presence of epidemic clones in a vulnerable population with contrasting characteristics and the presence of resistance genes call for strict vigilance.
Keywords: Escherichia coli; India; New Delhi metallo-β-lactamases; antibiotic resistance; epidemic clones; phylogroup; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Milton R, Gillespie D, Dyer C, Taiyari K, Carvalho MJ, Thomson K, Sands K, Portal EAR, Hood K, Ferreira A, Hender T, Kirby N, Mathias J, Nieto M, Watkins WJ, Bekele D, Abayneh M, Solomon S, Basu S, Nandy RK, Saha B, Iregbu K, Modibbo FZ, Uwaezuoke S, Zahra R, Shirazi H, Najeeb SU, Mazarati J-B, Rucogoza A, Gaju L, Mehtar S, Bulabula ANH, Whitelaw AC, Walsh TR, Chan GJ, BARNARDS Group . 2022. Neonatal sepsis and mortality in low-income and middle-income countries from a facility-based birth cohort: an international multisite prospective observational study. Lancet Glob Health 10:e661–72–e672. doi: 10.1016/S2214-109X(22)00043-2. - DOI - PMC - PubMed
-
- Gritz EC, Bhandari V. 2015. The human neonatal gut microbiome: a brief review. Front Pediatr 3. http://www.frontiersin.org/Neonatology/10.3389/fped.2015.00017/abstract. - DOI - PMC - PubMed
-
- Vila J, Sáez-López E, Johnson JR, Römling U, Dobrindt U, Cantón R, Giske CG, Naas T, Carattoli A, Martínez-Medina M, Bosch J, Retamar P, Rodríguez-Baño J, Baquero F, Soto SM. 2016. Escherichia coli: an old friend with new tidings. FEMS Microbiol Rev 40:437–463. doi: 10.1093/femsre/fuw005. - DOI - PubMed
-
- Mahjoub-Messai F, Bidet P, Caro V, Diancourt L, Biran V, Aujard Y, Bingen E, Bonacorsi S. 2011. Escherichia coli isolates causing bacteremia via gut translocation and urinary tract infection in young infants exhibit different virulence genotypes. J Infect Dis 203:1844–1849. doi: 10.1093/infdis/jir189. - DOI - PubMed
-
- Carl MA, Ndao IM, Springman AC, Manning SD, Johnson JR, Johnston BD, Burnham C-AD, Weinstock ES, Weinstock GM, Wylie TN, Mitreva M, Abubucker S, Zhou Y, Stevens HJ, Hall-Moore C, Julian S, Shaikh N, Warner BB, Tarr PI. 2014. Sepsis from the gut: the enteric habitat of bacteria that cause late-onset neonatal bloodstream infections. Clin Infect Dis 58:1211–1218. doi: 10.1093/cid/ciu084. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

