Antioxidant Peptides from Monkfish Swim Bladders: Ameliorating NAFLD In Vitro by Suppressing Lipid Accumulation and Oxidative Stress via Regulating AMPK/Nrf2 Pathway
- PMID: 37367685
- PMCID: PMC10301360
- DOI: 10.3390/md21060360
Antioxidant Peptides from Monkfish Swim Bladders: Ameliorating NAFLD In Vitro by Suppressing Lipid Accumulation and Oxidative Stress via Regulating AMPK/Nrf2 Pathway
Abstract
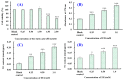
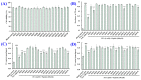
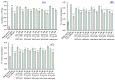
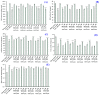
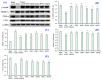
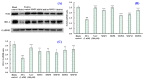
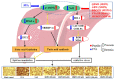
In this study, we investigate the ameliorating functions of QDYD (MSP2), ARW (MSP8), DDGGK (MSP10), YPAGP (MSP13) and DPAGP (MSP18) from monkfish swim bladders on an FFA-induced NAFLD model of HepG2 cells. The lipid-lowering mechanisms revealed that these five oligopeptides can up-regulate the expression of phospho-AMP-activated protein kinase (p-AMPK) proteins to inhibit the expression of the sterol regulatory element binding protein-1c (SREBP-1c) proteins on increasing lipid synthesis and up-regulating the expression of the PPAP-α and CPT-1 proteins on promoting the β-oxidation of fatty acids. Moreover, QDYD (MSP2), ARW (MSP8), DDGGK (MSP10), YPAGP (MSP13) and DPAGP (MSP18) can significantly inhibit reactive oxygen species' (ROS) production, promote the activities of intracellular antioxidases (superoxide dismutase, SOD; glutathione peroxidase, GSH-PX; and catalase, CAT) and bring down the content of malondialdehyde (MDA) derived from lipid peroxidation. Further investigations revealed that the regulation of these five oligopeptides on oxidative stress was achieved through activating the nuclear factor erythroid 2-related factor 2 (Nrf2) pathway to raise the expression levels of the heme oxygenase 1 (HO-1) protein and downstream antioxidant proteases. Therefore, QDYD (MSP2), ARW (MSP8), DDGGK (MSP10), YPAGP (MSP13) and DPAGP (MSP18) could serve as candidate ingredients to develop functional products for treating NAFLD.
Keywords: AMPK/Nrf2 pathway; antioxidant peptide; monkfish (Lophius litulon); nonalcoholic fatty liver disease (NAFLD); swim bladders.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

