An LRP1-binding motif in cellular prion protein replicates cell-signaling activities of the full-length protein
- PMID: 37368488
- PMCID: PMC10445690
- DOI: 10.1172/jci.insight.170121
An LRP1-binding motif in cellular prion protein replicates cell-signaling activities of the full-length protein
Abstract
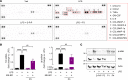
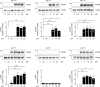
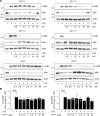
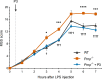
Low-density lipoprotein receptor-related protein-1 (LRP1) functions as a receptor for nonpathogenic cellular prion protein (PrPC), which is released from cells by ADAM (a disintegrin and metalloproteinase domain) proteases or in extracellular vesicles. This interaction activates cell signaling and attenuates inflammatory responses. We screened 14-mer PrPC-derived peptides and identified a putative LRP1 recognition motif in the PrPC sequence spanning residues 98-111. A synthetic peptide (P3) corresponding to this region replicated the cell-signaling and biological activities of full-length shed PrPC. P3 blocked LPS-elicited cytokine expression in macrophages and microglia and rescued the heightened sensitivity to LPS in mice in which the PrPC gene (Prnp) had been deleted. P3 activated ERK1/2 and induced neurite outgrowth in PC12 cells. The response to P3 required LRP1 and the NMDA receptor and was blocked by the PrPC-specific antibody, POM2. P3 has Lys residues, which are typically necessary for LRP1 binding. Converting Lys100 and Lys103 into Ala eliminated the activity of P3, suggesting that these residues are essential in the LRP1-binding motif. A P3 derivative in which Lys105 and Lys109 were converted into Ala retained activity. We conclude that the biological activities of shed PrPC, attributed to interaction with LRP1, are retained in synthetic peptides, which may be templates for therapeutics development.
Keywords: Cell Biology; Inflammation; Peptides; Prions; Signal transduction.
Conflict of interest statement
Figures


Similar articles
-
A soluble derivative of PrPC activates cell-signaling and regulates cell physiology through LRP1 and the NMDA receptor.J Biol Chem. 2020 Oct 9;295(41):14178-14188. doi: 10.1074/jbc.RA120.013779. Epub 2020 Aug 11. J Biol Chem. 2020. PMID: 32788217 Free PMC article.
-
Cellular prion protein in human plasma-derived extracellular vesicles promotes neurite outgrowth via the NMDA receptor-LRP1 receptor system.J Biol Chem. 2022 Mar;298(3):101642. doi: 10.1016/j.jbc.2022.101642. Epub 2022 Jan 25. J Biol Chem. 2022. PMID: 35090893 Free PMC article.
-
A Soluble PrPC Derivative and Membrane-Anchored PrPC in Extracellular Vesicles Attenuate Innate Immunity by Engaging the NMDA-R/LRP1 Receptor Complex.J Immunol. 2022 Jan 1;208(1):85-96. doi: 10.4049/jimmunol.2100412. Epub 2021 Nov 22. J Immunol. 2022. PMID: 34810220 Free PMC article.
-
Biology of the prion gene complex.Biochem Cell Biol. 2001;79(5):613-28. doi: 10.1139/o01-142. Biochem Cell Biol. 2001. PMID: 11716303 Review.
-
On the biology of prions.Acta Neuropathol. 1987;72(4):299-314. doi: 10.1007/BF00687261. Acta Neuropathol. 1987. PMID: 3554880 Review.
Cited by
-
CT1812 biomarker signature from a meta-analysis of CSF proteomic findings from two Phase 2 clinical trials in Alzheimer's disease.Alzheimers Dement. 2024 Oct;20(10):6860-6880. doi: 10.1002/alz.14152. Epub 2024 Aug 21. Alzheimers Dement. 2024. PMID: 39166791 Free PMC article. Clinical Trial.
-
Roles of prion proteins in mammalian development.Anim Cells Syst (Seoul). 2024 Dec 10;28(1):551-566. doi: 10.1080/19768354.2024.2436860. eCollection 2024. Anim Cells Syst (Seoul). 2024. PMID: 39664939 Free PMC article. Review.
-
Cleavage site-directed antibodies reveal the prion protein in humans is shed by ADAM10 at Y226 and associates with misfolded protein deposits in neurodegenerative diseases.Acta Neuropathol. 2024 Jul 9;148(1):2. doi: 10.1007/s00401-024-02763-5. Acta Neuropathol. 2024. PMID: 38980441 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

