Phenylalanine Residues in the Active Site of CYP2E1 Participate in Determining the Binding Orientation and Metabolism-Dependent Genotoxicity of Aromatic Compounds
- PMID: 37368596
- PMCID: PMC10302094
- DOI: 10.3390/toxics11060495
Phenylalanine Residues in the Active Site of CYP2E1 Participate in Determining the Binding Orientation and Metabolism-Dependent Genotoxicity of Aromatic Compounds
Abstract
The composition of amino acids forming the active site of a CYP enzyme is impactful in its substrate selectivity. For CYP2E1, the role of PHE residues in the formation of effective binding orientations for its aromatic substrates remains unclear. In this study, molecular docking and molecular dynamics analysis were performed to reflect the interactions between PHEs in the active site of human CYP2E1 and various aromatic compounds known as its substrates. The results indicated that the orientation of 1-methylpyrene (1-MP) in the active site was highly determined by the presence of PHEs, PHE478 contributing to the binding free energy most significantly. Moreover, by building a random forest model the relationship between each of 19 molecular descriptors of polychlorinated biphenyl (PCB) compounds (from molecular docking, quantum mechanics, and physicochemical properties) and their human CYP2E1-dependent mutagenicityas established mostly in our lab, was investigated. The presence of PHEs did not appear to significantly modify the electronic or structural feature of each bound ligand (PCB), instead, the flexibility of the conformation of PHEs contributed substantially to the effective binding energy and orientation. It is supposed that PHE residues adjust their own conformation to permit a suitablly shaped cavity for holding the ligand and forming its orientation as favorable for a biochemical reaction. This study has provided some insights into the role of PHEs in guiding the interactive adaptation of the active site of human CYP2E1 for the binding and metabolism of aromatic substrates.
Keywords: CYP2E1; aromatic compounds; molecular simulation; phenylalanine; random forest.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

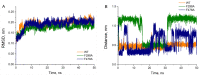

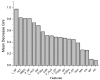
References
-
- Montellano PROd . Cytochrome P450 Structure, Mechanism, and Biochemistry. 4th ed. Springer International Publishing; Berlin/Heidelberg, Germany: 2015.
LinkOut - more resources
Full Text Sources
Miscellaneous

