Combining Nanopore Sequencing with Recombinase Polymerase Amplification Enables Identification of Dinoflagellates from the Alexandrium Genus, Providing a Rapid, Field Deployable Tool
- PMID: 37368673
- PMCID: PMC10302762
- DOI: 10.3390/toxins15060372
Combining Nanopore Sequencing with Recombinase Polymerase Amplification Enables Identification of Dinoflagellates from the Alexandrium Genus, Providing a Rapid, Field Deployable Tool
Abstract
The armoured dinoflagellate Alexandrium can be found throughout many of the world's temperate and tropical marine environments. The genus has been studied extensively since approximately half of its members produce a family of potent neurotoxins, collectively called saxitoxin. These compounds represent a significant threat to animal and environmental health. Moreover, the consumption of bivalve molluscs contaminated with saxitoxin poses a threat to human health. The identification of Alexandrium cells collected from sea water samples using light microscopy can provide early warnings of a toxic event, giving harvesters and competent authorities time to implement measures that safeguard consumers. However, this method cannot reliably resolve Alexandrium to a species level and, therefore, is unable to differentiate between toxic and non-toxic variants. The assay outlined in this study uses a quick recombinase polymerase amplification and nanopore sequencing method to first target and amplify a 500 bp fragment of the ribosomal RNA large subunit and then sequence the amplicon so that individual species from the Alexandrium genus can be resolved. The analytical sensitivity and specificity of the assay was assessed using seawater samples spiked with different Alexandrium species. When using a 0.22 µm membrane to capture and resuspend cells, the assay was consistently able to identify a single cell of A. minutum in 50 mL of seawater. Phylogenetic analysis showed the assay could identify the A. catenella, A. minutum, A. tamutum, A. tamarense, A. pacificum, and A. ostenfeldii species from environmental samples, with just the alignment of the reads being sufficient to provide accurate, real-time species identification. By using sequencing data to qualify when the toxic A. catenella species was present, it was possible to improve the correlation between cell counts and shellfish toxicity from r = 0.386 to r = 0.769 (p ≤ 0.05). Furthermore, a McNemar's paired test performed on qualitative data highlighted no statistical differences between samples confirmed positive or negative for toxic species of Alexandrium by both phylogenetic analysis and real time alignment with the presence or absence of toxins in shellfish. The assay was designed to be deployed in the field for the purposes of in situ testing, which required the development of custom tools and state-of-the-art automation. The assay is rapid and resilient to matrix inhibition, making it suitable as a potential alternative detection method or a complementary one, especially when applying regulatory controls.
Keywords: Alexandrium; HABs; RPA; VolTRAX; aquaculture; food safety; harmful algal bloom; in-field sequencing; nanopore sequencing; paralytic shellfish poisoning; saxitoxin.
Conflict of interest statement
The authors declare no conflict of interest, and the funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




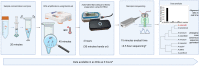



References
-
- Hallegraeff G.M. A review of harmful algal blooms and their apparent global increase. Phycologia. 1993;32:79–99. doi: 10.2216/i0031-8884-32-2-79.1. - DOI
-
- Hallegraeff G.M. Harmful algal blooms: A global overview. Man. Harmful Mar. Microalgae. 2003;33:1–22. doi: 10.1016/B978-0-12-391499-6.00008-6. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

