IL-7 receptor signaling drives human B-cell progenitor differentiation and expansion
- PMID: 37369082
- PMCID: PMC10644098
- DOI: 10.1182/blood.2023019721
IL-7 receptor signaling drives human B-cell progenitor differentiation and expansion
Abstract
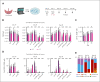
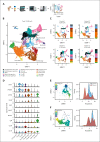
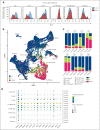
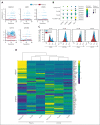
Although absence of interleukin-7 (IL-7) signaling completely abrogates T and B lymphopoiesis in mice, patients with severe combined immunodeficiency caused by mutations in the IL-7 receptor α chain (IL-7Rα) still generate peripheral blood B cells. Consequently, human B lymphopoiesis has been thought to be independent of IL-7 signaling. Using flow cytometric analysis and single-cell RNA sequencing of bone marrow samples from healthy controls and patients who are IL-7Rα deficient, in combination with in vitro modeling of human B-cell differentiation, we demonstrate that IL-7R signaling plays a crucial role in human B lymphopoiesis. IL-7 drives proliferation and expansion of early B-cell progenitors but not of pre-BII large cells and has a limited role in the prevention of cell death. Furthermore, IL-7 guides cell fate decisions by enhancing the expression of BACH2, EBF1, and PAX5, which jointly orchestrate the specification and commitment of early B-cell progenitors. In line with this observation, early B-cell progenitors of patients with IL-7Rα deficiency still expressed myeloid-specific genes. Collectively, our results unveil a previously unknown role for IL-7 signaling in promoting the B-lymphoid fate and expanding early human B-cell progenitors while defining important differences between mice and humans. Our results have implications for hematopoietic stem cell transplantation strategies in patients with T- B+ severe combined immunodeficiency and provide insights into the role of IL-7R signaling in leukemogenesis.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures






Comment in
-
The "B" side of IL-7Rα-deficient SCID.Blood. 2023 Sep 28;142(13):1105-1106. doi: 10.1182/blood.2023021475. Blood. 2023. PMID: 37768696 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

