Self-patterning of human stem cells into post-implantation lineages
- PMID: 37369348
- PMCID: PMC10584676
- DOI: 10.1038/s41586-023-06354-4
Self-patterning of human stem cells into post-implantation lineages
Abstract
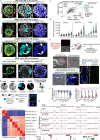
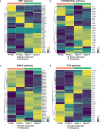
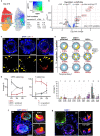
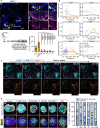
Investigating human development is a substantial scientific challenge due to the technical and ethical limitations of working with embryonic samples. In the face of these difficulties, stem cells have provided an alternative to experimentally model inaccessible stages of human development in vitro1-13. Here we show that human pluripotent stem cells can be triggered to self-organize into three-dimensional structures that recapitulate some key spatiotemporal events of early human post-implantation embryonic development. Our system reproducibly captures spontaneous differentiation and co-development of embryonic epiblast-like and extra-embryonic hypoblast-like lineages, establishes key signalling hubs with secreted modulators and undergoes symmetry breaking-like events. Single-cell transcriptomics confirms differentiation into diverse cell states of the perigastrulating human embryo14,15 without establishing placental cell types, including signatures of post-implantation epiblast, amniotic ectoderm, primitive streak, mesoderm, early extra-embryonic endoderm, as well as initial yolk sac induction. Collectively, our system captures key features of human embryonic development spanning from Carnegie stage16 4-7, offering a reproducible, tractable and scalable experimental platform to understand the basic cellular and molecular mechanisms that underlie human development, including new opportunities to dissect congenital pathologies with high throughput.
© 2023. The Author(s).
Conflict of interest statement
All authors declare no competing interests.
Figures










Comment in
-
Dawn of development: Exploring early human embryogenesis using stem cells.Cell Stem Cell. 2023 Aug 3;30(8):1006-1007. doi: 10.1016/j.stem.2023.07.009. Cell Stem Cell. 2023. PMID: 37541206
-
Models of the early human embryo.Nat Biotechnol. 2023 Aug;41(8):1072. doi: 10.1038/s41587-023-01920-7. Nat Biotechnol. 2023. PMID: 37568022 No abstract available.
-
Stem cells used to model a two-week-old human embryo.Nature. 2023 Oct;622(7983):469-470. doi: 10.1038/d41586-023-03150-y. Nature. 2023. PMID: 37848524 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

