A New Breakpoint to Classify 3D Voxels in MRI: A Space Transform Strategy with 3t2FTS-v2 and Its Application for ResNet50-Based Categorization of Brain Tumors
- PMID: 37370560
- PMCID: PMC10294956
- DOI: 10.3390/bioengineering10060629
A New Breakpoint to Classify 3D Voxels in MRI: A Space Transform Strategy with 3t2FTS-v2 and Its Application for ResNet50-Based Categorization of Brain Tumors
Abstract
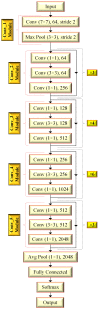
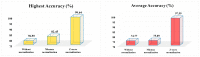
Three-dimensional (3D) image analyses are frequently applied to perform classification tasks. Herein, 3D-based machine learning systems are generally used/generated by examining two designs: a 3D-based deep learning model or a 3D-based task-specific framework. However, except for a new approach named 3t2FTS, a promising feature transform operating from 3D to two-dimensional (2D) space has not been efficiently investigated for classification applications in 3D magnetic resonance imaging (3D MRI). In other words, a state-of-the-art feature transform strategy is not available that achieves high accuracy and provides the adaptation of 2D-based deep learning models for 3D MRI-based classification. With this aim, this paper presents a new version of the 3t2FTS approach (3t2FTS-v2) to apply a transfer learning model for tumor categorization of 3D MRI data. For performance evaluation, the BraTS 2017/2018 dataset is handled that involves high-grade glioma (HGG) and low-grade glioma (LGG) samples in four different sequences/phases. 3t2FTS-v2 is proposed to effectively transform the features from 3D to 2D space by using two textural features: first-order statistics (FOS) and gray level run length matrix (GLRLM). In 3t2FTS-v2, normalization analyses are assessed to be different from 3t2FTS to accurately transform the space information apart from the usage of GLRLM features. The ResNet50 architecture is preferred to fulfill the HGG/LGG classification due to its remarkable performance in tumor grading. As a result, for the classification of 3D data, the proposed model achieves a 99.64% accuracy by guiding the literature about the importance of 3t2FTS-v2 that can be utilized not only for tumor grading but also for whole brain tissue-based disease classification.
Keywords: brain; convolutional neural network; dimensional; feature transform; glioma grading; image classification; transfer learning; tumor.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
Similar articles
-
Deep Multi-Scale 3D Convolutional Neural Network (CNN) for MRI Gliomas Brain Tumor Classification.J Digit Imaging. 2020 Aug;33(4):903-915. doi: 10.1007/s10278-020-00347-9. J Digit Imaging. 2020. PMID: 32440926 Free PMC article.
-
A comprehensive study of brain tumour discrimination using phase combinations, feature rankings, and hybridised classifiers.Med Biol Eng Comput. 2020 Dec;58(12):2971-2987. doi: 10.1007/s11517-020-02273-y. Epub 2020 Oct 2. Med Biol Eng Comput. 2020. PMID: 33006703
-
Learning Architecture for Brain Tumor Classification Based on Deep Convolutional Neural Network: Classic and ResNet50.Diagnostics (Basel). 2025 Mar 5;15(5):624. doi: 10.3390/diagnostics15050624. Diagnostics (Basel). 2025. PMID: 40075870 Free PMC article.
-
Convolutional neural networks for classification of Alzheimer's disease: Overview and reproducible evaluation.Med Image Anal. 2020 Jul;63:101694. doi: 10.1016/j.media.2020.101694. Epub 2020 May 1. Med Image Anal. 2020. PMID: 32417716
-
Machine learning applications to neuroimaging for glioma detection and classification: An artificial intelligence augmented systematic review.J Clin Neurosci. 2021 Jul;89:177-198. doi: 10.1016/j.jocn.2021.04.043. Epub 2021 May 13. J Clin Neurosci. 2021. PMID: 34119265
References
-
- AboElenein N.M., Piao S., Noor A., Ahmed P.N. MIRAU-Net: An improved neural network based on U-Net for gliomas segmentation. Signal Process.-Image. 2022;101:116553. doi: 10.1016/j.image.2021.116553. - DOI
-
- Hu Z., Li L., Sui A., Wu G., Wang Y., Yu J. An efficient R-Transformer network with dual encoders for brain glioma segmentation in MR images. Biomed. Signal Proces. 2023;79:104034. doi: 10.1016/j.bspc.2022.104034. - DOI
-
- Gumaei A., Hassan M.M., Hassan M.R., Alelaiwi A., Fortino G. A hybrid feature extraction method with regularized extreme learning machine for brain tumor classification. IEEE Access. 2019;7:36266–36273. doi: 10.1109/ACCESS.2019.2904145. - DOI
-
- Alshayeji M., Al-Buloushi J., Ashkanani A., Abed S.E. Enhanced brain tumor classification using an optimized multi-layered convolutional neural network architecture. Multimed. Tools Appl. 2021;80:28897–28917. doi: 10.1007/s11042-021-10927-8. - DOI
LinkOut - more resources
Full Text Sources
Research Materials