Transcriptomic Analysis Reveals Candidate Ligand-Receptor Pairs and Signaling Networks Mediating Intercellular Communication between Hair Matrix Cells and Dermal Papilla Cells from Cashmere Goats
- PMID: 37371115
- PMCID: PMC10297330
- DOI: 10.3390/cells12121645
Transcriptomic Analysis Reveals Candidate Ligand-Receptor Pairs and Signaling Networks Mediating Intercellular Communication between Hair Matrix Cells and Dermal Papilla Cells from Cashmere Goats
Abstract
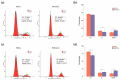
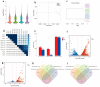
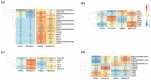
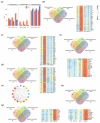
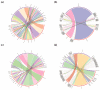
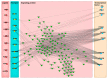
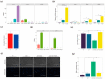
Hair fiber growth is determined by the spatiotemporally controlled proliferation, differentiation, and apoptosis of hair matrix cells (HMCs) inside the hair follicle (HF); however, dermal papilla cells (DPCs), the cell population surrounded by HMCs, manipulate the above processes via intercellular crosstalk with HMCs. Therefore, exploring how the mutual commutations between the cells are molecularly achieved is vital to understanding the mechanisms underlying hair growth. Here, based on our previous successes in cultivating HMCs and DPCs from cashmere goats, we combined a series of techniques, including in vitro cell coculture, transcriptome sequencing, and bioinformatic analysis, to uncover ligand-receptor pairs and signaling networks mediating intercellular crosstalk. Firstly, we found that direct cellular interaction significantly alters cell cycle distribution patterns and changes the gene expression profiles of both cells at the global level. Next, we constructed the networks of ligand-receptor pairs mediating intercellular autocrine or paracrine crosstalk between the cells. A few pairs, such as LEP-LEPR, IL6-EGFR, RSPO1-LRP6, and ADM-CALCRL, are found to have known or potential roles in hair growth by acting as bridges linking cells. Further, we inferred the signaling axis connecting the cells from transcriptomic data with the advantage of CCCExplorer. Certain pathways, including INHBA-ACVR2A/ACVR2B-ACVR1/ACVR1B-SMAD3, were predicted as the axis mediating the promotive effect of INHBA on hair growth via paracrine crosstalk between DPCs and HMCs. Finally, we verified that LEP-LEPR and IL1A-IL1R1 are pivotal ligand-receptor pairs involved in autocrine and paracrine communication of DPCs and HMCs to DPCs, respectively. Our study provides a comprehensive landscape of intercellular crosstalk between key cell types inside HF at the molecular level, which is helpful for an in-depth understanding of the mechanisms related to hair growth.
Keywords: cashmere goat; dermal papilla cells; hair follicle; hair matrix cells; intercellular crosstalk; ligand-receptor pair; signaling axis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

