Leaflet Tensions Control the Spatio-Temporal Remodeling of Lipid Bilayers and Nanovesicles
- PMID: 37371505
- PMCID: PMC10296112
- DOI: 10.3390/biom13060926
Leaflet Tensions Control the Spatio-Temporal Remodeling of Lipid Bilayers and Nanovesicles
Abstract
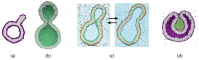
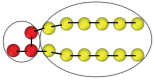
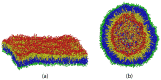
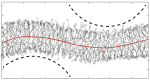
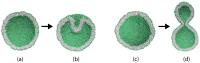
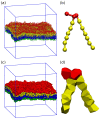
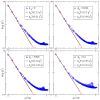
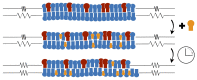
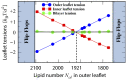
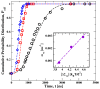
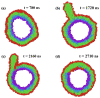
Biological and biomimetic membranes are based on lipid bilayers, which consist of two monolayers or leaflets. To avoid bilayer edges, which form when the hydrophobic core of such a bilayer is exposed to the surrounding aqueous solution, a single bilayer closes up into a unilamellar vesicle, thereby separating an interior from an exterior aqueous compartment. Synthetic nanovesicles with a size below 100 nanometers, traditionally called small unilamellar vesicles, have emerged as potent platforms for the delivery of drugs and vaccines. Cellular nanovesicles of a similar size are released from almost every type of living cell. The nanovesicle morphology has been studied by electron microscopy methods but these methods are limited to a single snapshot of each vesicle. Here, we review recent results of molecular dynamics simulations, by which one can monitor and elucidate the spatio-temporal remodeling of individual bilayers and nanovesicles. We emphasize the new concept of leaflet tensions, which control the bilayers' stability and instability, the transition rates of lipid flip-flops between the two leaflets, the shape transformations of nanovesicles, the engulfment and endocytosis of condensate droplets and rigid nanoparticles, as well as nanovesicle adhesion and fusion. To actually compute the leaflet tensions, one has to determine the bilayer's midsurface, which represents the average position of the interface between the two leaflets. Two particularly useful methods to determine this midsurface are based on the density profile of the hydrophobic lipid chains and on the molecular volumes.
Keywords: bilayer tension; biomembranes; endocytosis; engulfment; leaflet tensions; molecular dynamics; simulations; synthetic biosystems.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
























Similar articles
-
Probing the elastic response of lipid bilayers and nanovesicles to leaflet tensions via volume per lipid.Soft Matter. 2023 Sep 20;19(36):6929-6944. doi: 10.1039/d3sm00351e. Soft Matter. 2023. PMID: 37664906
-
Large stress asymmetries of lipid bilayers and nanovesicles generate lipid flip-flops and bilayer instabilities.Soft Matter. 2022 Aug 17;18(32):6066-6078. doi: 10.1039/d2sm00618a. Soft Matter. 2022. PMID: 35929498
-
Multiscale remodeling of biomembranes and vesicles.Methods Enzymol. 2024;701:175-236. doi: 10.1016/bs.mie.2024.04.006. Epub 2024 May 9. Methods Enzymol. 2024. PMID: 39025572
-
Remodeling of Biomembranes and Vesicles by Adhesion of Condensate Droplets.Membranes (Basel). 2023 Feb 10;13(2):223. doi: 10.3390/membranes13020223. Membranes (Basel). 2023. PMID: 36837726 Free PMC article. Review.
-
Do sterols reduce proton and sodium leaks through lipid bilayers?Prog Lipid Res. 2001 Jul;40(4):299-324. doi: 10.1016/s0163-7827(01)00009-1. Prog Lipid Res. 2001. PMID: 11412894 Review.
Cited by
-
Inhomogeneous Canham-Helfrich Abscission in Catenoid Necks under Critical Membrane Mosaicity.Membranes (Basel). 2023 Sep 14;13(9):796. doi: 10.3390/membranes13090796. Membranes (Basel). 2023. PMID: 37755218 Free PMC article.
-
Membrane Association of Intrinsically Disordered Proteins.Annu Rev Biophys. 2025 May;54(1):275-302. doi: 10.1146/annurev-biophys-070124-092816. Epub 2025 Feb 14. Annu Rev Biophys. 2025. PMID: 39952269 Review.
-
Multispherical shapes of vesicles with intramembrane domains.Eur Phys J E Soft Matter. 2024 Jan 11;47(1):4. doi: 10.1140/epje/s10189-023-00399-z. Eur Phys J E Soft Matter. 2024. PMID: 38206459 Free PMC article.
References
-
- Robertson J.D. The ultrastructure of cell membranes and their derivatives. Biochem. Soc. Symp. 1959;16:3–43. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

