Real-Time Search-Assisted Multiplexed Quantitative Proteomics Reveals System-Wide Translational Regulation of Non-Canonical Short Open Reading Frames
- PMID: 37371559
- PMCID: PMC10296652
- DOI: 10.3390/biom13060979
Real-Time Search-Assisted Multiplexed Quantitative Proteomics Reveals System-Wide Translational Regulation of Non-Canonical Short Open Reading Frames
Abstract
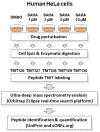
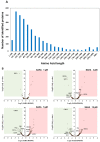
Abnormal expression of histone deacetylases (HDACs) is reported to be associated with angiogenesis, metastasis and chemotherapy resistance regarding cancer in a wide range of previous studies. Suberoylanilide hydroxamic acid (SAHA) is well known to function as a pan-inhibitor for HDACs and recognized as one of the therapeutic drug candidates to epigenetically coordinate cancer cell fate regulation on a genomic scale. Here, we established a Real-Time Search (RTS)-assisted mass spectrometric platform for system-wide quantification of translated products encoded by non-canonical short open reading frames (ORFs) as well as already annotated protein coding sequences (CDSs) on the human transciptome and applied this methodology to quantitative proteomic analyses of suberoylanilide hydroxamic acid (SAHA)-treated human HeLa cells to evaluate proteome-wide regulation in response to drug perturbation. Very intriguingly, our RTS-based in-depth proteomic analysis enabled us to identify approximately 5000 novel peptides from the ribosome profiling-based short ORFs encoded in the diversified regions on presumed 'non-coding' nucleotide sequences of mRNAs as well as lncRNAs and nonsense mediated decay (NMD) transcripts. Furthermore, TMT-based multiplex large-scale quantification of the whole proteome changes upon differential SAHA treatment unveiled dose-dependent selective translational regulation of a limited fraction of the non-canonical short ORFs in addition to key cell cycle/proliferation-related molecules such as UBE2C, CENPF and PRC1. Our study provided the first system-wide landscape of drug-perturbed translational modulation on both canonical and non-canonical proteome dynamics in human cancer cells.
Keywords: cancer; histone deacetylase; proteomics; real-time search; short open reading frames.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

