Pre- and Post-Transcriptional Control of HBV Gene Expression: The Road Traveled towards the New Paradigm of HBx, Its Isoforms, and Their Diverse Functions
- PMID: 37371770
- PMCID: PMC10296101
- DOI: 10.3390/biomedicines11061674
Pre- and Post-Transcriptional Control of HBV Gene Expression: The Road Traveled towards the New Paradigm of HBx, Its Isoforms, and Their Diverse Functions
Abstract
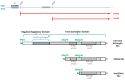
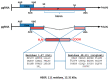
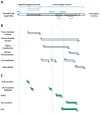
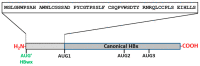
Hepatitis B virus (HBV) is an enveloped DNA human virus belonging to the Hepadnaviridae family. Perhaps its main distinguishable characteristic is the replication of its genome through a reverse transcription process. The HBV circular genome encodes only four overlapping reading frames, encoding for the main canonical proteins named core, P, surface, and X (or HBx protein). However, pre- and post-transcriptional gene regulation diversifies the full HBV proteome into diverse isoform proteins. In line with this, hepatitis B virus X protein (HBx) is a viral multifunctional and regulatory protein of 16.5 kDa, whose canonical reading frame presents two phylogenetically conserved internal in-frame translational initiation codons, and which results as well in the expression of two divergent N-terminal smaller isoforms of 8.6 and 5.8 kDa, during translation. The canonical HBx, as well as the smaller isoform proteins, displays different roles during viral replication and subcellular localizations. In this article, we reviewed the different mechanisms of pre- and post-transcriptional regulation of protein expression that take place during viral replication. We also investigated all the past and recent evidence about HBV HBx gene regulation and its divergent N-terminal isoform proteins. Evidence has been collected for over 30 years. The accumulated evidence simply strengthens the concept of a new paradigm of the canonical HBx, and its smaller divergent N-terminal isoform proteins, not only during viral replication, but also throughout cell pathogenesis.
Keywords: HBx; hepatitis B virus; hepatitis B virus X protein; isoform protein; mRNA; pre- and post-transcriptional control; viral regulatory protein; viral replication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

