Polygenic Variants Linked to Oxidative Stress and the Antioxidant System Are Associated with Type 2 Diabetes Risk and Interact with Lifestyle Factors
- PMID: 37372010
- PMCID: PMC10295348
- DOI: 10.3390/antiox12061280
Polygenic Variants Linked to Oxidative Stress and the Antioxidant System Are Associated with Type 2 Diabetes Risk and Interact with Lifestyle Factors
Abstract
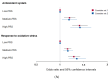
Oxidative stress is associated with insulin resistance and secretion, and antioxidant systems are essential for preventing and managing type 2 diabetes (T2DM). This study aimed to explore the polygenic variants linked to oxidative stress and the antioxidant system among those associated with T2DM and the interaction of their polygenic risk scores (PRSs) with lifestyle factors in a large hospital-based cohort (n = 58,701). Genotyping, anthropometric, biochemical, and dietary assessments were conducted for all participants with an average body mass index of 23.9 kg/m2. Genetic variants associated with T2DM were searched through genome-wide association studies in participants with T2DM (n = 5383) and without T2DM (n = 53,318). The Gene Ontology database was searched for the antioxidant systems and oxidative stress-related genes among the genetic variants associated with T2DM risk, and the PRS was generated by summing the risk alleles of selected ones. Gene expression according to the genetic variant alleles was determined on the FUMA website. Food components with low binding energy to the GSTA5 protein generated from the wildtype and mutated GSTA5_rs7739421 (missense mutation) genes were selected using in silico analysis. Glutathione metabolism-related genes, including glutathione peroxidase (GPX)1 and GPX3, glutathione disulfide reductase (GSR), peroxiredoxin-6 (PRDX6), glutamate-cysteine ligase catalytic subunit (GCLC), glutathione S-transferase alpha-5 (GSTA5), and gamma-glutamyltransferase-1 (GGT1), were predominantly selected with a relevance score of >7. The PRS related to the antioxidant system was positively associated with T2DM (ORs = 1.423, 95% CI = 1.22-1.66). The active site of the GASTA proteins having valine or leucine at 55 due to the missense mutation (rs7739421) had a low binding energy (<-10 kcal/mol) similarly or differently to some flavonoids and anthocyanins. The PRS interacted with the intake of bioactive components (specifically dietary antioxidants, vitamin C, vitamin D, and coffee) and smoking status (p < 0.05). In conclusion, individuals with a higher PRS related to the antioxidant system may have an increased risk of T2DM, and there is a potential indication that exogenous antioxidant intake may alleviate this risk, providing insights for personalized strategies in T2DM prevention.
Keywords: antioxidants; bioactive compounds; coffee; oxidative stress; type 2 diabetes; vitamin D.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Association of Polygenic Variants with Type 2 Diabetes Risk and Their Interaction with Lifestyles in Asians.Nutrients. 2022 Aug 6;14(15):3222. doi: 10.3390/nu14153222. Nutrients. 2022. PMID: 35956399 Free PMC article.
-
Interaction of Polygenetic Variants for Gestational Diabetes Mellitus Risk with Breastfeeding and Korean Balanced Diet to Influence Type 2 Diabetes Risk in Later Life in a Large Hospital-Based Cohort.J Pers Med. 2021 Nov 10;11(11):1175. doi: 10.3390/jpm11111175. J Pers Med. 2021. PMID: 34834527 Free PMC article.
-
Gene-Lifestyle Interactions in Renal Dysfunction: Polygenic Risk Modulation via Plant-Based Diets, Coffee Intake, and Bioactive Compound Interactions.Nutrients. 2025 Mar 6;17(5):916. doi: 10.3390/nu17050916. Nutrients. 2025. PMID: 40077791 Free PMC article.
-
Dietary glycation compounds - implications for human health.Crit Rev Toxicol. 2024 Sep;54(8):485-617. doi: 10.1080/10408444.2024.2362985. Epub 2024 Aug 16. Crit Rev Toxicol. 2024. PMID: 39150724
-
The thioredoxin antioxidant system.Free Radic Biol Med. 2014 Jan;66:75-87. doi: 10.1016/j.freeradbiomed.2013.07.036. Epub 2013 Jul 27. Free Radic Biol Med. 2014. PMID: 23899494 Review.
Cited by
-
Genotype-based precision nutrition strategies for the prediction and clinical management of type 2 diabetes mellitus.World J Diabetes. 2024 Feb 15;15(2):142-153. doi: 10.4239/wjd.v15.i2.142. World J Diabetes. 2024. PMID: 38464367 Free PMC article. Review.
-
Associations of dietary selenium intake with the risk of chronic diseases and mortality in US adults.Front Nutr. 2024 Jun 24;11:1363299. doi: 10.3389/fnut.2024.1363299. eCollection 2024. Front Nutr. 2024. PMID: 38978702 Free PMC article.
-
Tryptophan Indole Derivatives: Key Players in Type 2 Diabetes Mellitus.Diabetes Metab Syndr Obes. 2025 May 12;18:1563-1574. doi: 10.2147/DMSO.S511068. eCollection 2025. Diabetes Metab Syndr Obes. 2025. PMID: 40386349 Free PMC article. Review.
-
The Role of Glutathione and Its Precursors in Type 2 Diabetes.Antioxidants (Basel). 2024 Feb 1;13(2):184. doi: 10.3390/antiox13020184. Antioxidants (Basel). 2024. PMID: 38397782 Free PMC article. Review.
-
Ethnicity-related differences in mitochondrial regulation by insulin stimulation in diabetes.J Cell Physiol. 2024 Aug;239(8):e31317. doi: 10.1002/jcp.31317. Epub 2024 May 22. J Cell Physiol. 2024. PMID: 38775168 Free PMC article. Review.
References
-
- Sun H., Saeedi P., Karuranga S., Pinkepank M., Ogurtsova K., Duncan B.B., Stein C., Basit A., Chan J.C.N., Mbanya J.C., et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022;183:109119. doi: 10.1016/j.diabres.2021.109119. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

