Compositional Structure of the Genome: A Review
- PMID: 37372134
- PMCID: PMC10295253
- DOI: 10.3390/biology12060849
Compositional Structure of the Genome: A Review
Abstract
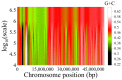
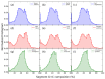
As the genome carries the historical information of a species' biotic and environmental interactions, analyzing changes in genome structure over time by using powerful statistical physics methods (such as entropic segmentation algorithms, fluctuation analysis in DNA walks, or measures of compositional complexity) provides valuable insights into genome evolution. Nucleotide frequencies tend to vary along the DNA chain, resulting in a hierarchically patchy chromosome structure with heterogeneities at different length scales that range from a few nucleotides to tens of millions of them. Fluctuation analysis reveals that these compositional structures can be classified into three main categories: (1) short-range heterogeneities (below a few kilobase pairs (Kbp)) primarily attributed to the alternation of coding and noncoding regions, interspersed or tandem repeats densities, etc.; (2) isochores, spanning tens to hundreds of tens of Kbp; and (3) superstructures, reaching sizes of tens of megabase pairs (Mbp) or even larger. The obtained isochore and superstructure coordinates in the first complete T2T human sequence are now shared in a public database. In this way, interested researchers can use T2T isochore data, as well as the annotations for different genome elements, to check a specific hypothesis about genome structure. Similarly to other levels of biological organization, a hierarchical compositional structure is prevalent in the genome. Once the compositional structure of a genome is identified, various measures can be derived to quantify the heterogeneity of such structure. The distribution of segment G+C content has recently been proposed as a new genome signature that proves to be useful for comparing complete genomes. Another meaningful measure is the sequence compositional complexity (SCC), which has been used for genome structure comparisons. Lastly, we review the recent genome comparisons in species of the ancient phylum Cyanobacteria, conducted by phylogenetic regression of SCC against time, which have revealed positive trends towards higher genome complexity. These findings provide the first evidence for a driven progressive evolution of genome compositional structure.
Keywords: DNA compositional structure; evolutionary adaptive trends; hierarchical genome structure; segment compositional signature; sequence compositional complexity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Bernardi G. Structural and Evolutionary Genomics: Natural Selection in Genome Evolution. Elsevier; Amsterdam, The Netherlands: 2004.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

