Genetic Diversity of Durum Wheat (Triticum turgidum L. ssp. durum, Desf) Germplasm as Revealed by Morphological and SSR Markers
- PMID: 37372335
- PMCID: PMC10298645
- DOI: 10.3390/genes14061155
Genetic Diversity of Durum Wheat (Triticum turgidum L. ssp. durum, Desf) Germplasm as Revealed by Morphological and SSR Markers
Abstract
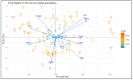
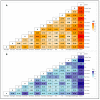
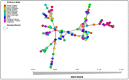
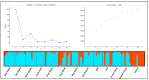
Ethiopia is considered a center of origin and diversity for durum wheat and is endowed with many diverse landraces. This research aimed to estimate the extent and pattern of genetic diversity in Ethiopian durum wheat germplasm. Thus, 104 durum wheat genotypes representing thirteen populations, three regions, and four altitudinal classes were investigated for their genetic diversity, using 10 grain quality- and grain yield-related phenotypic traits and 14 simple sequence repeat (SSR) makers. The analysis of the phenotypic traits revealed a high mean Shannon diversity index (H' = 0.78) among the genotypes and indicated a high level of phenotypic variation. The principal component analysis (PCA) classified the genotypes into three groups. The SSR markers showed a high mean value of polymorphic information content (PIC = 0.50) and gene diversity (h = 0.56), and a moderate number of alleles per locus (Na = 4). Analysis of molecular variance (AMOVA) revealed a high level of variation within populations, regions, and altitudinal classes, accounting for 88%, 97%, and 97% of the total variation, respectively. Pairwise genetic differentiation and Nei's genetic distance analyses identified that the cultivars are distinct from the landrace populations. The distance-based (Discriminant Analysis of Principal Component (DAPC) and Minimum Spanning Network (MSN)) and model-based population stratification (STRUCTURE) methods of clustering grouped the genotypes into two clusters. Both the phenotypic data-based PCA and the molecular data-based DAPC and MSN analyses defined distinct groupings of cultivars and landraces. The phenotypic and molecular diversity analyses highlighted the high genetic variation in the Ethiopian durum wheat gene pool. The investigated SSRs showed significant associations with one or more target phenotypic traits. The markers identify landraces with high grain yield and quality traits. This study highlights the usefulness of Ethiopian landraces for cultivar development, contributing to food security in the region and beyond.
Keywords: SSR markers; durum wheat; genetic diversity; landraces; morphological traits.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Hancock J.F. Plant Evolution and the Origin of Crop Species. 2nd ed. Volume 123. CABI Publishing; Cambridge, UK: 2004.
-
- Özkan H., Willcox G., Graner A., Salamini F., Kilian B. Geographic distribution and domestication of wild emmer wheat (Triticum dicoccoides) Genet. Resour. Crop. Evol. 2011;58:11–53. doi: 10.1007/s10722-010-9581-5. - DOI
-
- Pecetti L., Annicchiarico P., Damania A.B. Biodiversity in a germplasm collection of durum wheat. Euphytica. 1992;60:229–238. doi: 10.1007/BF00039403. - DOI
-
- Kabbaj H., Sall A.T., Al-Abdallat A., Geleta M., Amri A., Filali-Maltouf A., Belkadi B., Ortiz R., Bassi F.M. Genetic Diversity within a Global Panel of Durum Wheat (Triticum durum) Landraces and Modern Germplasm Reveals the History of Alleles Exchange. Front. Plant Sci. 2017;8:1277. doi: 10.3389/fpls.2017.01277. - DOI - PMC - PubMed
-
- Teklu Y., Hammer K., Huang X., Röder M. Analysis of Microsatellite Diversity in Ethiopian Tetraploid Wheat Landraces. Genet. Resour. Crop Evol. 2006;53:1115–1126. doi: 10.1007/s10722-005-1146-7. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

