Genetic Diversity and Population Structure in Türkiye Bread Wheat Genotypes Revealed by Simple Sequence Repeats (SSR) Markers
- PMID: 37372362
- PMCID: PMC10298624
- DOI: 10.3390/genes14061182
Genetic Diversity and Population Structure in Türkiye Bread Wheat Genotypes Revealed by Simple Sequence Repeats (SSR) Markers
Abstract
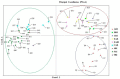
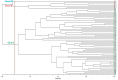
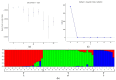
Wheat genotypes should be improved through available germplasm genetic diversity to ensure food security. This study investigated the molecular diversity and population structure of a set of Türkiye bread wheat genotypes using 120 microsatellite markers. Based on the results, 651 polymorphic alleles were evaluated to determine genetic diversity and population structure. The number of alleles ranged from 2 to 19, with an average of 5.44 alleles per locus. Polymorphic information content (PIC) ranged from 0.031 to 0.915 with a mean of 0.43. In addition, the gene diversity index ranged from 0.03 to 0.92 with an average of 0.46. The expected heterozygosity ranged from 0.00 to 0.359 with a mean of 0.124. The unbiased expected heterozygosity ranged from 0.00 to 0.319 with an average of 0.112. The mean values of the number of effective alleles (Ne), genetic diversity of Nei (H) and Shannon's information index (I) were estimated at 1.190, 1.049 and 0.168, respectively. The highest genetic diversity (GD) was estimated between genotypes G1 and G27. In the UPGMA dendrogram, the 63 genotypes were grouped into three clusters. The three main coordinates were able to explain 12.64, 6.38 and 4.90% of genetic diversity, respectively. AMOVA revealed diversity within populations at 78% and between populations at 22%. The current populations were found to be highly structured. Model-based cluster analyses classified the 63 genotypes studied into three subpopulations. The values of F-statistic (Fst) for the identified subpopulations were 0.253, 0.330 and 0.244, respectively. In addition, the expected values of heterozygosity (He) for these sub-populations were recorded as 0.45, 0.46 and 0.44, respectively. Therefore, SSR markers can be useful not only in genetic diversity and association analysis of wheat but also in its germplasm for various agronomic traits or mechanisms of tolerance to environmental stresses.
Keywords: SSR; bread wheat; gene diversity; population structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Horvath A., Didier A., Koenig J., Exbrayat F., Charmet G., Balfourier F. Analysis of diversity and linkage disequilibrium along chromosome 3B of bread wheat (Triticum aestivum L.) Theor. Appl. Genet. 2009;119:1523–1537. - PubMed
-
- Dvorak J., McGuire P.E., Cassidy B. Apparent sources of the A genomes of wheats inferred from polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome. 1988;30:680–689. doi: 10.1139/g88-115. - DOI
-
- Savadi S., Prasad P., Kashyap P., Bhardwaj S. Molecular breeding technologies and strategies for rust resistance in wheat (Triticum aestivum) for sustained food security. Plant Pathol. 2018;67:771–791. doi: 10.1111/ppa.12802. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

