Determination of Effect Sizes for Power Analysis for Microbiome Studies Using Large Microbiome Databases
- PMID: 37372419
- PMCID: PMC10297957
- DOI: 10.3390/genes14061239
Determination of Effect Sizes for Power Analysis for Microbiome Studies Using Large Microbiome Databases
Abstract
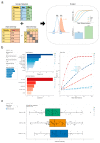
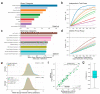
Herein, we present a tool called Evident that can be used for deriving effect sizes for a broad spectrum of metadata variables, such as mode of birth, antibiotics, socioeconomics, etc., to provide power calculations for a new study. Evident can be used to mine existing databases of large microbiome studies (such as the American Gut Project, FINRISK, and TEDDY) to analyze the effect sizes for planning future microbiome studies via power analysis. For each metavariable, the Evident software is flexible to compute effect sizes for many commonly used measures of microbiome analyses, including α diversity, β diversity, and log-ratio analysis. In this work, we describe why effect size and power analysis are necessary for computational microbiome analysis and show how Evident can help researchers perform these procedures. Additionally, we describe how Evident is easy for researchers to use and provide an example of efficient analyses using a dataset of thousands of samples and dozens of metadata categories.
Keywords: bioinformatics; effect size; microbiome; statistics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Cohen J. Statistical Power Analysis. Curr. Dir. Psychol. Sci. 1992;1:98–101. doi: 10.1111/1467-8721.ep10768783. - DOI

