Incorporation of N7-Platinated Guanines into Thermus Aquaticus (Taq) DNA Polymerase: Atomistic Insights from Molecular Dynamics Simulations
- PMID: 37372996
- PMCID: PMC10297917
- DOI: 10.3390/ijms24129849
Incorporation of N7-Platinated Guanines into Thermus Aquaticus (Taq) DNA Polymerase: Atomistic Insights from Molecular Dynamics Simulations
Abstract
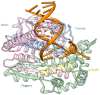
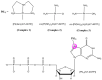
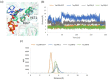
In this work, we elucidated some key aspects of the mechanism of action of the cisplatin anticancer drug, cis-[Pt(NH3)2Cl2], involving direct interactions with free nucleotides. A comprehensive in silico molecular modeling analysis was conducted to compare the interactions of Thermus aquaticus (Taq) DNA polymerase with three distinct N7-platinated deoxyguanosine triphosphates: [Pt(dien)(N7-dGTP)] (1), cis-[Pt(NH3)2Cl(N7-dGTP)] (2), and cis-[Pt(NH3)2(H2O)(N7-dGTP)] (3) {dien = diethylenetriamine; dGTP = 5'-(2'-deoxy)-guanosine-triphosphate}, using canonical dGTP as a reference, in the presence of DNA. The goal was to elucidate the binding site interactions between Taq DNA polymerase and the tested nucleotide derivatives, providing valuable atomistic insights. Unbiased molecular dynamics simulations (200 ns for each complex) with explicit water molecules were performed on the four ternary complexes, yielding significant findings that contribute to a better understanding of experimental results. The molecular modeling highlighted the crucial role of a specific α-helix (O-helix) within the fingers subdomain, which facilitates the proper geometry for functional contacts between the incoming nucleotide and the DNA template needed for incorporation into the polymerase. The analysis revealed that complex 1 exhibits a much lower affinity for Taq DNA polymerase than complexes 2-3. The affinities of cisplatin metabolites 2-3 for Taq DNA polymerase were found to be quite similar to those of natural dGTP, resulting in a lower incorporation rate for complex 1 compared to complexes 2-3. These findings could have significant implications for the cisplatin mechanism of action, as the high intracellular availability of free nucleobases might promote the competitive incorporation of platinated nucleotides over direct cisplatin attachment to DNA. The study's insights into the incorporation of platinated nucleotides into the Taq DNA polymerase active site suggest that the role of platinated nucleotides in the cisplatin mechanism of action may have been previously underestimated.
Keywords: antimetabolites; antitumor drugs; antiviral drugs; cisplatin; coordination compounds; molecular dynamics; nucleoside analogues; platinum compounds.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


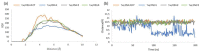

References
-
- Anthony E.J., Bolitho E.M., Bridgewater H.E., Carter O.W.L., Donnelly J.M., Imberti C., Lant E.C., Lermyte F., Needham R.J., Palau M., et al. Metallodrugs Are Unique: Opportunities and Challenges of Discovery and Development. Chem. Sci. 2020;11:12888–12917. doi: 10.1039/D0SC04082G. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

