Etiologies of Acute Bronchiolitis in Children at Risk for Asthma, with Emphasis on the Human Rhinovirus Genotyping Protocol
- PMID: 37373604
- PMCID: PMC10299285
- DOI: 10.3390/jcm12123909
Etiologies of Acute Bronchiolitis in Children at Risk for Asthma, with Emphasis on the Human Rhinovirus Genotyping Protocol
Abstract
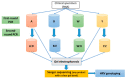
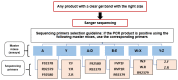
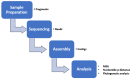
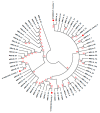
This research aims to determine acute bronchiolitis' causative virus(es) and establish a viable protocol to classify the Human Rhinovirus (HRV) species. During 2021-2022, we included children 1-24 months of age with acute bronchiolitis at risk for asthma. The nasopharyngeal samples were taken and subjected to a quantitative polymerase chain reaction (qPCR) in a viral panel. For HRV-positive samples, a high-throughput assay was applied, directing the VP4/VP2 and VP3/VP1 regions to confirm species. BLAST searching, phylogenetic analysis, and sequence divergence took place to identify the degree to which these regions were appropriate for identifying and differentiating HRV. HRV ranked second, following RSV, as the etiology of acute bronchiolitis in children. The conclusion of the investigation of all available data in this study distributed sequences into 7 HRV-A, 1 HRV-B, and 7 HRV-C types based on the VP4/VP2 and VP3/VP1 sequences. The nucleotide divergence between the clinical samples and the corresponding reference strains was lower in the VP4/VP2 region than in the VP3/VP1 region. The results demonstrated the potential utility of the VP4/VP2 region and the VP3/VP1 region for differentiating HRV genotypes. Confirmatory outcomes were yielded, indicating how nested and semi-nested PCR can establish practical ways to facilitate HRV sequencing and genotyping.
Keywords: acute bronchiolitis; asthma; bioinformatics; genotyping; human rhinovirus; sequencing; virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Jartti T., Lehtinen P., Vuorinen T., Österback R., van den Hoogen B., Osterhaus A.D., Ruuskanen O. Respiratory picornaviruses and respiratory syncytial virus as causative agents of acute expiratory wheezing in children. Emerg. Infect. Dis. 2004;10:1095–1101. doi: 10.3201/eid1006.030629. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

