Synthesis, Characterization, DFT Studies of Novel Cu(II), Zn(II), VO(II), Cr(III), and La(III) Chloro-Substituted Schiff Base Complexes: Aspects of Its Antimicrobial, Antioxidant, Anti-Inflammatory, and Photodegradation of Methylene Blue
- PMID: 37375332
- PMCID: PMC10301192
- DOI: 10.3390/molecules28124777
Synthesis, Characterization, DFT Studies of Novel Cu(II), Zn(II), VO(II), Cr(III), and La(III) Chloro-Substituted Schiff Base Complexes: Aspects of Its Antimicrobial, Antioxidant, Anti-Inflammatory, and Photodegradation of Methylene Blue
Abstract
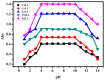
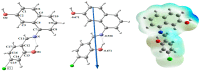
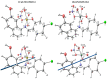
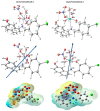
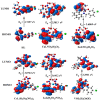
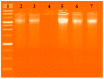
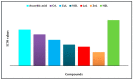
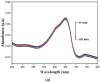
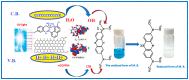
A new chlorobenzylidene imine ligand, (E)-1-((5-chloro-2-hydroxybenzylidene)amino) naphthalen-2-ol (HL), and its [Zn(L)(NO3)(H2O)3], [La(L)(NO3)2(H2O)2], [VO(L)(OC2H5)(H2O)2], [Cu(L)(NO3)(H2O)3], and [Cr(L)(NO3)2(H2O)2], complexes were synthesized and characterized. The characterization involved elemental analysis, FT-IR, UV/Vis, NMR, mass spectra, molar conductance, and magnetic susceptibility measurements. The obtained data confirmed the octahedral geometrical structures of all metal complexes, while the [VO(L)(OC2H5)(H2O)2] complex exhibited a distorted square pyramidal structure. The complexes were found to be thermally stable based on their kinetic parameters determined using the Coats-Redfern method. The DFT/B3LYP technique was employed to calculate the optimized structures, energy gaps, and other important theoretical descriptors of the complexes. In vitro antibacterial assays were conducted to evaluate the complexes' potential against pathogenic bacteria and fungi, comparing them to the free ligand. The compounds exhibited excellent fungicidal activity against Candida albicans ATCC: 10231 (C. albicans) and Aspergillus negar ATCC: 16404 (A. negar), with inhibition zones of HL, [Zn(L)(NO3)(H2O)3], and [La(L)(NO3)2(H2O)2] three times higher than that of the Nystatin antibiotic. The DNA binding affinity of the metal complexes and their ligand was investigated using UV-visible, viscosity, and gel electrophoresis methods, suggesting an intercalative binding mode. The absorption studies yielded Kb values ranging from 4.40 × 105 to 7.30 × 105 M-1, indicating high binding strength to DNA comparable to ethidium bromide (value 107 M-1). Additionally, the antioxidant activity of all complexes was measured and compared to vitamin C. The anti-inflammatory efficacy of the ligand and its metal complexes was evaluated, revealing that [Cu(L)(NO3)(H2O)3] exhibited the most effective activity compared to ibuprofen. Molecular docking studies were conducted to explore the binding nature and affinity of the synthesized compounds with the receptor of Candida albicans oxidoreductase/oxidoreductase INHIBITOR (PDB ID: 5V5Z). Overall, the combined findings of this work demonstrate the potential of these new compounds as efficient fungicidal and anti-inflammatory agents. Furthermore, the photocatalytic effect of the Cu(II) Schiff base complex/GO was examined.
Keywords: DFT; DNA-binding; Schiff bases; anti-inflammatory; antimicrobial; antioxidant; photodegradation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures













References
-
- Abd El-Halim H.F.A., Mohamed G.G., Khalil E.A. Synthesis, spectral, thermal and biological studies of mixed ligand complexes with newly prepared Schiff base and 1,10-phenanthroline ligands. J. Mol. Struct. 2017;1146:153–163. doi: 10.1016/j.molstruc.2017.05.092. - DOI
-
- Mohamed R.G., Elantabli F.M., Helal N.H., El-Medani S.M. New group 6 metal carbonyl complexes with 4,5-dimethyl-N,N-bis(pyridine-2-yl-methylene)benzene-1,2-diimine Schiff base: Synthesis, spectral, cyclic voltammetry and biological activity studies. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015;141:316–326. doi: 10.1016/j.saa.2015.01.054. - DOI - PubMed
-
- Ali O.A.M. Palladium(II) and zinc(II) complexes of neutral [N2O2] donor Schiff bases derived from furfuraldehyde: Synthesis, characterization, fluorescence and corrosion inhibitors of ligands. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014;132:52–60. doi: 10.1016/j.saa.2014.03.127. - DOI - PubMed
-
- Nasrin D., Alam M.A., Hossain M.N., Nazimuddin M. Synthesis, Characterization and Antimicrobial Activity of Metal Complexes of Schiff ’ s Base Derived from S-benzyldithiocarbazate with 2-hydroxyacetophenone. Chem. J. 2013;3:13–19.
-
- Sherif O.E., Abdel-Kader N.S. Spectroscopic and biological activities studies of bivalent transition metal complexes of Schiff bases derived from condensation of 1,4-phenylenediamine and benzopyrone derivatives. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014;117:519–526. doi: 10.1016/j.saa.2013.08.037. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

