CLEAR Strategy Inhibited HSV Proliferation Using Viral Vectors Delivered CRISPR-Cas9
- PMID: 37375504
- PMCID: PMC10302303
- DOI: 10.3390/pathogens12060814
CLEAR Strategy Inhibited HSV Proliferation Using Viral Vectors Delivered CRISPR-Cas9
Abstract
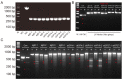
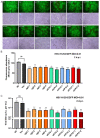
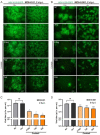
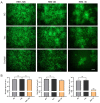
Herpes simplex virus type 1 (HSV-1) is a leading cause of encephalitis and infectious blindness. The commonly used clinical therapeutic drugs are nucleoside analogues such as acyclovir. However, current drugs for HSV cannot eliminate the latent virus or viral reactivation. Therefore, the development of new treatment strategies against latent HSV has become an urgent need. To comprehensively suppress the proliferation of HSV, we designed the CLEAR strategy (coordinated lifecycle elimination against viral replication). VP16, ICP27, ICP4, and gD-which are crucial genes that perform significant functions in different stages of the HSV infection lifecycle-were selected as targeting sites based on CRISPR-Cas9 editing system. In vitro and in vivo investigations revealed that genome editing by VP16, ICP27, ICP4 or gD single gene targeting could effectively inhibit HSV replication. Moreover, the combined administration method (termed "Cocktail") showed superior effects compared to single gene editing, which resulted in the greatest decrease in viral proliferation. Lentivirus-delivered CRISPR-Cas9/gRNA editing could effectively block HSV replication. The CLEAR strategy may provide new insights into the potential treatment of refractory HSV-1-associated diseases, particularly when conventional approaches have encountered resistance.
Keywords: CRISPR-Cas9; ICP27; ICP4; VP16; gD; gRNA; herpes simplex virus; lentivirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Suppression of HSV-1 infection and viral reactivation by CRISPR-Cas9 gene editing in 2D and 3D culture models.Mol Ther Nucleic Acids. 2024 Jul 19;35(3):102282. doi: 10.1016/j.omtn.2024.102282. eCollection 2024 Sep 10. Mol Ther Nucleic Acids. 2024. PMID: 39176174 Free PMC article.
-
Single AAV-Mediated CRISPR-SaCas9 Inhibits HSV-1 Replication by Editing ICP4 in Trigeminal Ganglion Neurons.Mol Ther Methods Clin Dev. 2020 May 22;18:33-43. doi: 10.1016/j.omtm.2020.05.011. eCollection 2020 Sep 11. Mol Ther Methods Clin Dev. 2020. PMID: 32577430 Free PMC article.
-
A knockdown of the herpes simplex virus type-1 gene in all-in-one CRISPR vectors.Folia Histochem Cytobiol. 2020;58(3):174-181. doi: 10.5603/FHC.a2020.0020. Epub 2020 Sep 16. Folia Histochem Cytobiol. 2020. PMID: 32937678
-
Development of Genome Editing Approaches against Herpes Simplex Virus Infections.Viruses. 2021 Feb 22;13(2):338. doi: 10.3390/v13020338. Viruses. 2021. PMID: 33671590 Free PMC article. Review.
-
Clinical management of herpes simplex virus infections: past, present, and future.F1000Res. 2018 Oct 31;7:F1000 Faculty Rev-1726. doi: 10.12688/f1000research.16157.1. eCollection 2018. F1000Res. 2018. PMID: 30443341 Free PMC article. Review.
Cited by
-
Applying CRISPR Technologies for the Treatment of Human Herpesvirus Infections: A Scoping Review.Pathogens. 2025 Jul 1;14(7):654. doi: 10.3390/pathogens14070654. Pathogens. 2025. PMID: 40732701 Free PMC article. Review.
-
Control of HSV-1 Infection: Directions for the Development of CRISPR/Cas-Based Therapeutics and Diagnostics.Int J Mol Sci. 2024 Nov 17;25(22):12346. doi: 10.3390/ijms252212346. Int J Mol Sci. 2024. PMID: 39596412 Free PMC article. Review.
-
The opportunities and challenges of epigenetic approaches to manage herpes simplex infections.Expert Rev Anti Infect Ther. 2024 Dec;22(12):1123-1142. doi: 10.1080/14787210.2024.2420329. Epub 2024 Nov 6. Expert Rev Anti Infect Ther. 2024. PMID: 39466139 Review.
-
CRISPR/Cas9 Eye Drop HSV-1 Treatment Reduces Brain Viral Load: A Novel Application to Prevent Neuronal Damage.Pathogens. 2024 Dec 10;13(12):1087. doi: 10.3390/pathogens13121087. Pathogens. 2024. PMID: 39770347 Free PMC article.
-
CRISPR-Cas9-mediated genome editing delivered by a single AAV9 vector inhibits HSV-1 reactivation in a latent rabbit keratitis model.Mol Ther Methods Clin Dev. 2024 Aug 14;32(3):101303. doi: 10.1016/j.omtm.2024.101303. eCollection 2024 Sep 12. Mol Ther Methods Clin Dev. 2024. PMID: 39610766 Free PMC article.
References
Grants and funding
- 2021ZD0201003/STI2030-Major Projects
- JCYJ20210324101413037/Shenzhen Science and Technology Program
- 31771198/National Natural Science Foundation of China
- 31830035/National Natural Science Foundation of China
- XDB32030200/Strategic Priority Research Program of the Chinese Academy of Sciences
- ZDSYS20200811142401005/Shenzhen Key Laboratory of Viral Vectors for Biomedicine
- JCYJ20220818100800001/Science, Technology and Innovation Commission of Shenzhen
- 2023ZDZ08/Key Laboratory of Quality Control Technology for Virus-Based Therapeutics, Guangdong Provin-cial Medical Products Administration
- 2022ZDZ13/Key Laboratory of Quality Control Technology for Virus-Based Therapeutics, Guangdong Provin-cial Medical Products Administration
LinkOut - more resources
Full Text Sources
Research Materials

