Upregulation of Neuropilin-1 Inhibits HTLV-1 Infection
- PMID: 37375521
- PMCID: PMC10305173
- DOI: 10.3390/pathogens12060831
Upregulation of Neuropilin-1 Inhibits HTLV-1 Infection
Abstract
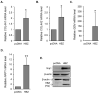
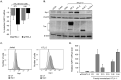
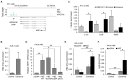
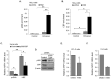
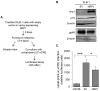
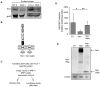
Infection with human T-cell leukemia virus type 1 (HTLV-1) can produce a spectrum of pathological effects ranging from inflammatory disorders to leukemia. In vivo, HTLV-1 predominantly infects CD4+ T-cells. Infectious spread within this population involves the transfer of HTLV-1 virus particles from infected cells to target cells only upon cell-to-cell contact. The viral protein, HBZ, was found to enhance HTLV-1 infection through transcriptional activation of ICAM1 and MYOF, two genes that facilitate viral infection. In this study, we found that HBZ upregulates the transcription of COL4A1, GEM, and NRP1. COL4A1 and GEM are genes involved in viral infection, while NRP1, which encodes neuropilin 1 (Nrp1), serves as an HTLV-1 receptor on target cells but has no reported function on HTLV-1-infected cells. With a focus on Nrp1, cumulative results from chromatin immunoprecipitation assays and analyses of HBZ mutants support a model in which HBZ upregulates NRP1 transcription by augmenting recruitment of Jun proteins to an enhancer downstream of the gene. Results from in vitro infection assays demonstrate that Nrp1 expressed on HTLV-1-infected cells inhibits viral infection. Nrp1 was found to be incorporated into HTLV-1 virions, and deletion of its ectodomain removed the inhibitory effect. These results suggest that inhibition of HTLV-1 infection by Nrp1 is caused by the ectodomain of Nrp1 extended from virus particles, which may inhibit the binding of virus particles to target cells. While HBZ has been found to enhance HTLV-1 infection using cell-based models, there may be certain circumstances in which activation of Nrp1 expression negatively impacts viral infection, which is discussed.
Keywords: HBZ; HTLV-1; Nrp1; bZIP; infection; p300/CBP; transcription.
Conflict of interest statement
The authors declare no conflict of interest. The funder had no role in the design of this study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

