Genomic Epidemiology of SARS-CoV-2 in Urban Settings in Senegal
- PMID: 37376533
- PMCID: PMC10302768
- DOI: 10.3390/v15061233
Genomic Epidemiology of SARS-CoV-2 in Urban Settings in Senegal
Abstract
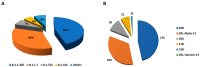
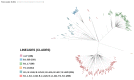
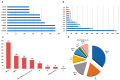
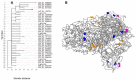
We used whole genome sequencing to identify and analyze mutations in SARS-CoV-2 in urban settings during the deadliest wave of the COVID-19 epidemic-from March to April 2021-in Senegal. Nasopharyngeal samples testing positive for SARS-CoV-2 were sequenced on the Illumina NovaSeq 6000 sequencing system using the COVIDSeq protocol. A total of 291 genotypable consensus genome sequences were obtained. Phylogenetic analyses grouped the genomes into 16 distinct PANGOLIN lineages. The major lineage was B.1.1.420, despite circulation of the Alpha variant of concern (VOC). A total of 1125 different SNPs, identified relative to the Wuhan reference genome, were detected. These included 13 SNPs in non-coding regions. An average density of 37.2 SNPs per 1000 nucleotides was found, with the highest density occurring in ORF10. This analysis allowed, for the first time, the detection of a Senegalese SARS-CoV-2 strain belonging to the P.1.14 (GR/20J, Gamma V3) sublineage of the Brazilian P.1 lineage (or Gamma VOC). Overall, our results highlight substantial SARS-CoV-2 diversification in Senegal during the study period.
Keywords: COVIDSeq; SARS-CoV-2; SNPs; Senegal; epidemiology; genomic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- World Health Organization Middle East Respiratory Syndrome Coronavirus (MERS-CoV) [(accessed on 19 May 2022)]. Available online: https://www.who.int/news-room/fact-sheets/detail/middle-east-respiratory...
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

