Multiple Lineages of Hantaviruses Harbored by the Iberian Mole (Talpa occidentalis) in Spain
- PMID: 37376613
- PMCID: PMC10302183
- DOI: 10.3390/v15061313
Multiple Lineages of Hantaviruses Harbored by the Iberian Mole (Talpa occidentalis) in Spain
Abstract
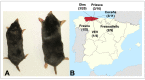
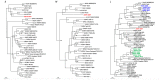
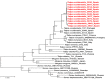
The recent detection of both Nova virus (NVAV) and Bruges virus (BRGV) in European moles (Talpa europaea) in Belgium and Germany prompted a search for related hantaviruses in the Iberian mole (Talpa occidentalis). RNAlater®-preserved lung tissue from 106 Iberian moles, collected during January 2011 to June 2014 in Asturias, Spain, were analyzed for hantavirus RNA by nested/hemi-nested RT-PCR. Pairwise alignment and comparison of partial L-segment sequences, detected in 11 Iberian moles from four parishes, indicated the circulation of genetically distinct hantaviruses. Phylogenetic analyses, using maximum-likelihood and Bayesian methods, demonstrated three distinct hantaviruses in Iberian moles: NVAV, BRGV, and a new hantavirus, designated Asturias virus (ASTV). Of the cDNA from seven infected moles processed for next generation sequencing using Illumina HiSeq1500, one produced viable contigs, spanning the S, M and L segments of ASTV. The original view that each hantavirus species is harbored by a single small-mammal host species is now known to be invalid. Host-switching or cross-species transmission events, as well as reassortment, have shaped the complex evolutionary history and phylogeography of hantaviruses such that some hantavirus species are hosted by multiple reservoir species, and conversely, some host species harbor more than one hantavirus species.
Keywords: Hantaviridae; Spain; evolution; hantavirus; talpid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Brummer-Korvenkontio M., Vaheri A., Hovi T., von Bonsdorff C.H., Vuorimies J., Manni T., Penttinen K., Oker-Blom N., Lähdevirta J. Nephropathia epidemica: Detection of antigen in bank voles and serologic diagnosis of human infection. J. Infect. Dis. 1980;141:131–134. doi: 10.1093/infdis/141.2.131. - DOI - PubMed
-
- Duchin J.S., Koster F.T., Peters C.J., Simpson G.L., Tempest B., Zaki S.R., Ksiazek T.G., Rollin P.E., Nichol S., Umland E.T., et al. Hantavirus pulmonary syndrome: A clinical description of 17 patients with a newly recognized disease. N. Engl. J. Med. 1994;330:949–955. doi: 10.1056/NEJM199404073301401. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

