A Novel Simian Adenovirus Associating with Human Adeno-virus Species G Isolated from Long-Tailed Macaque Feces
- PMID: 37376670
- PMCID: PMC10303043
- DOI: 10.3390/v15061371
A Novel Simian Adenovirus Associating with Human Adeno-virus Species G Isolated from Long-Tailed Macaque Feces
Erratum in
-
Correction: Kosoltanapiwat et al. A Novel Simian Adenovirus Associating with Human Adenovirus Species G Isolated from Long-Tailed Macaque Feces. Viruses 2023, 15, 1371.Viruses. 2023 Sep 4;15(9):1871. doi: 10.3390/v15091871. Viruses. 2023. PMID: 37766380 Free PMC article.
Abstract
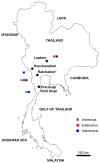
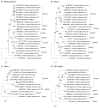
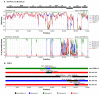
Metagenomics has demonstrated its capability in outbreak investigations and pathogen surveillance and discovery. With high-throughput and effective bioinformatics, many disease-causing agents, as well as novel viruses of humans and animals, have been identified using metagenomic analysis. In this study, a VIDISCA metagenomics workflow was used to identify potential unknown viruses in 33 fecal samples from asymptomatic long-tailed macaques (Macaca fascicularis) in Ratchaburi Province, Thailand. Putatively novel astroviruses, enteroviruses, and adenoviruses were detected and confirmed by PCR analysis of long-tailed macaque fecal samples collected from areas in four provinces, Ratchaburi, Kanchanaburi, Lopburi, and Prachuap Khiri Khan, where humans and monkeys live in proximity (total n = 187). Astroviruses, enteroviruses, and adenoviruses were present in 3.2%, 7.5%, and 4.8% of macaque fecal samples, respectively. One adenovirus, named AdV-RBR-6-3, was successfully isolated in human cell culture. Whole-genome analysis suggested that it is a new member of the species Human adenovirus G, closely related to Rhesus adenovirus 53, with evidence of genetic recombination and variation in the hexon, fiber, and CR1 genes. Sero-surveillance showed neutralizing antibodies against AdV-RBR-6-3 in 2.9% and 11.2% of monkeys and humans, respectively, suggesting cross-species infection of monkeys and humans. Overall, we reported the use of metagenomics to screen for possible new viruses, as well as the isolation and molecular and serological characterization of the new adenovirus with cross-species transmission potential. The findings emphasize that zoonotic surveillance is important and should be continued, especially in areas where humans and animals interact, to predict and prevent the threat of emerging zoonotic pathogens.
Keywords: VIDISCA; long-tailed macaques; metagenomics; novel adenovirus; virus discovery.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


Similar articles
-
Simian adenoviruses: Molecular and serological survey in monkeys and humans in Thailand.One Health. 2022 Sep 16;15:100434. doi: 10.1016/j.onehlt.2022.100434. eCollection 2022 Dec. One Health. 2022. PMID: 36277107 Free PMC article.
-
Isolation of a recombinant simian adenovirus encoding the human adenovirus G52 hexon suggests a simian origin for human adenovirus G52.J Virol. 2024 Apr 16;98(4):e0004324. doi: 10.1128/jvi.00043-24. Epub 2024 Mar 18. J Virol. 2024. PMID: 38497664 Free PMC article.
-
High Diversity and Novel Enteric Viruses in Fecal Viromes of Healthy Wild and Captive Thai Cynomolgus Macaques (Macaca fascicularis).Viruses. 2019 Oct 22;11(10):971. doi: 10.3390/v11100971. Viruses. 2019. PMID: 31652508 Free PMC article.
-
A first report of non-invasive adenovirus detection in wild Assamese macaques in Thailand.Primates. 2017 Apr;58(2):307-313. doi: 10.1007/s10329-016-0587-2. Epub 2016 Nov 17. Primates. 2017. PMID: 27858173
-
Is the long-tailed macaque at risk of extinction?Am J Primatol. 2024 Apr;86(4):e23590. doi: 10.1002/ajp.23590. Epub 2023 Dec 21. Am J Primatol. 2024. PMID: 38124676 Review.
Cited by
-
Correction: Kosoltanapiwat et al. A Novel Simian Adenovirus Associating with Human Adenovirus Species G Isolated from Long-Tailed Macaque Feces. Viruses 2023, 15, 1371.Viruses. 2023 Sep 4;15(9):1871. doi: 10.3390/v15091871. Viruses. 2023. PMID: 37766380 Free PMC article.
-
Adenovirus infections: new insights for the clinical laboratory.J Clin Microbiol. 2024 Sep 11;62(9):e0083622. doi: 10.1128/jcm.00836-22. Epub 2024 Aug 27. J Clin Microbiol. 2024. PMID: 39189703 Free PMC article. Review.
-
Novel Mastadenovirus Infection as Cause of Pneumonia in Imported Black-and-White Colobuses (Colobus guereza), Thailand.Emerg Infect Dis. 2024 Dec;30(12):2544-2558. doi: 10.3201/eid3012.241042. Emerg Infect Dis. 2024. PMID: 39592267 Free PMC article.
References
-
- Suwannarong K., Soonthornworasiri N., Maneekan P., Balthip K., Yimsamran S., Maneewatchararangsri S., Ponlap T., Saengkul C., Lantican C., Thammasutti K., et al. Love or conflict: A qualitative study of the human-long tailed macaque interface in Nakhon Sawan Province, Thailand. Acta Trop. 2023;240:106861. doi: 10.1016/j.actatropica.2023.106861. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

